Difference between revisions of "Voxel Meshing Module"
From NAMIC Wiki
| (12 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
**[[Coordinate System Conversion Between ITK and Slicer3]] | **[[Coordinate System Conversion Between ITK and Slicer3]] | ||
* Extraction of surfaces nodes has been implemented to view results in Slicer3 | * Extraction of surfaces nodes has been implemented to view results in Slicer3 | ||
| + | * Material properties can be assigned to the elements using the Voxel data signal intensity and converting this into a modulus value. The user can specify the number of unique values to use for the modulus. | ||
| + | **Rebinning can be done based on the minimum and maximum value and then divide this into equal steps between these values | ||
| + | **A user specified histogram can also be done using a user specified histogram | ||
| + | *Output formats | ||
| + | **VTK | ||
| + | **[http://www.simulia.com/ Abaqus] | ||
| + | * Source code updated to support VTK 5.0 instead of VTK 5.1. This change required that the histogram bin data is written as Field Data in a VTK file. | ||
| + | * Meshing module is now built by default with Slicer3 | ||
| + | * This code was updated to integrate with the new meshing module. This should be available in Slicer3.4 release | ||
| − | |||
| − | |||
'''Key Investigators:''' | '''Key Investigators:''' | ||
| Line 18: | Line 25: | ||
'''Links:''' | '''Links:''' | ||
| − | *[http://www.ccad.uiowa.edu/mimx Musculoskeletal Imaging, | + | *[http://www.ccad.uiowa.edu/mimx Musculoskeletal Imaging, Modeling and Experimentation (MIMX)] |
| + | |||
| + | '''Description:''' | ||
| + | This program will convert an image into a hexahedral mesh using | ||
| + | the voxels as the hexahedral elements. This is is typically done | ||
| + | for only a portion of the image and thus a binary image from a | ||
| + | segmentation may be used to define the region for meshing. | ||
| + | Since these meshes may be fairly large the user is allowed | ||
| + | to downsample the image by an integer value. Currently this | ||
| + | must be odd. | ||
| + | |||
| + | This tool was developed as part of an NIH NA-MIC Collaboration Grant EB005973. | ||
| + | |||
| + | '''Program Usage:''' | ||
| + | VoxelMeshingModule | ||
| + | |||
| + | --imagefilename Input image File Name | ||
| + | --maskfilename Input mask File Name | ||
| + | --abaqusfilename Output Abaqus Filename | ||
| + | --vtkfilename Output mesh VTK filename | ||
| + | --vtksurfacefilename Output mesh surface VTK filename (Hack required for slicer3) | ||
| + | --resamplesize The image resample downsampling factor - Must be odd | ||
| + | --meshindexoffset The mesh index offset for nodes and elements | ||
| + | --poissonratio Poisson Ratio for the mesh | ||
| + | --abaqustitle Abaqus title for generated mesh | ||
| + | --numberofbins The number of histogram bins in the mesh | ||
| + | --histogamlowerbound The lower bound for the mesh histogram | ||
| + | --histogamupperbound The upper bound for the mesh histogram | ||
| + | --histogambinfile Mesh material properties histogram bin filename | ||
| + | |||
| + | '''Figures:''' | ||
| + | |||
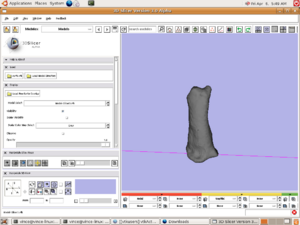
| + | [[Image:Slicer3Surface.png|left|thumb|300px|Surface of proximal phalanx generated via Slicer3]] | ||
| + | |||
| + | [[Image:VoxelMesh.png|left|thumb|300px|Voxel mesh generated via Slicer3 execution Model]] | ||
| + | |||
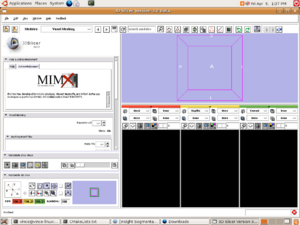
| + | [[Image:Mimx.png|left|thumb|300px|Slicer3 Voxel Mesh Module]] | ||
Latest revision as of 21:05, 9 December 2008
Home < Voxel Meshing ModuleObjective:
- Develop an initial framework for integration of Meshing modules into the NA-MIC kit
Progress:
- Base code has been developed as an Execution Model for Slicer3
- Orientatation issues wrt Slicer3 and ITK coordinate systems have been determined
- Extraction of surfaces nodes has been implemented to view results in Slicer3
- Material properties can be assigned to the elements using the Voxel data signal intensity and converting this into a modulus value. The user can specify the number of unique values to use for the modulus.
- Rebinning can be done based on the minimum and maximum value and then divide this into equal steps between these values
- A user specified histogram can also be done using a user specified histogram
- Output formats
- VTK
- Abaqus
- Source code updated to support VTK 5.0 instead of VTK 5.1. This change required that the histogram bin data is written as Field Data in a VTK file.
- Meshing module is now built by default with Slicer3
- This code was updated to integrate with the new meshing module. This should be available in Slicer3.4 release
Key Investigators:
- Iowa: Nicole Grosland, Vincent Magnotta, Ritesh Bafna
Links:
Description: This program will convert an image into a hexahedral mesh using the voxels as the hexahedral elements. This is is typically done for only a portion of the image and thus a binary image from a segmentation may be used to define the region for meshing. Since these meshes may be fairly large the user is allowed to downsample the image by an integer value. Currently this must be odd.
This tool was developed as part of an NIH NA-MIC Collaboration Grant EB005973.
Program Usage:
VoxelMeshingModule --imagefilename Input image File Name --maskfilename Input mask File Name --abaqusfilename Output Abaqus Filename --vtkfilename Output mesh VTK filename --vtksurfacefilename Output mesh surface VTK filename (Hack required for slicer3) --resamplesize The image resample downsampling factor - Must be odd --meshindexoffset The mesh index offset for nodes and elements --poissonratio Poisson Ratio for the mesh --abaqustitle Abaqus title for generated mesh --numberofbins The number of histogram bins in the mesh --histogamlowerbound The lower bound for the mesh histogram --histogamupperbound The upper bound for the mesh histogram --histogambinfile Mesh material properties histogram bin filename
Figures: