Difference between revisions of "2014 Summer Project Week:Atlas Selection"
| (8 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
==Key Investigators== | ==Key Investigators== | ||
* Kanglin Chen (Fraunhofer MEVIS Germany) | * Kanglin Chen (Fraunhofer MEVIS Germany) | ||
| − | * Gregory Sharp ( | + | * Gregory Sharp (MGH) |
==Project Description== | ==Project Description== | ||
| Line 14: | Line 14: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
Atlas selection is used for image segmentation. Normally, a single image is chosen as an atlas and the structures are segmented manually. | Atlas selection is used for image segmentation. Normally, a single image is chosen as an atlas and the structures are segmented manually. | ||
| − | The segmentation is transferred to patient data using non-linear image registration. However, image segmentation based on single atlas is | + | The segmentation is transferred to patient data using non-linear image registration. However, image segmentation based on single image atlas is |
| − | not stable. To improve the segmentation we can select multiple images, which are suitable for image segmentation | + | not stable. To improve the segmentation, we can select multiple images, which are suitable for image segmentation. After registration we |
| − | can merge the segmentations to a final one. | + | can merge the segmentations to a final one. An idea to improve atlas selection process lies on that we take advantage of using an average atlas. |
| − | * Construct an atlas based on a database | + | The procedure of atlas selection using an average atlas for image segmentation is defined as follows: |
| − | * Every image of the database is aligned to the atlas after atlas construction | + | * Construct an average atlas based on a database |
| − | * Register the atlas to a new image and transfer the database to the new image | + | * Every image of the database is aligned to the average atlas after the atlas construction. |
| − | * Compare the transformed database | + | * Register the average atlas to a new image and transfer the database and their segmentations to the new image. |
| − | * Merge the segmentations of these images to a final one based on e.g. | + | * Compare the transformed database with the new image in the ROI and select some well-matched images with respected to some distance measure, e.g. MI, CC, NGF. |
| − | The crucial step of atlas selection is atlas construction | + | * Merge the segmentations of these well-matched images to a final one based on e.g. weighted voting or STAPLE algorithm. |
| + | The crucial step of atlas selection is the average atlas construction. The focus of this project lies on the construction of an average atlas using 3D datasets and validate the average atlas with the inspection of segmentations. | ||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | The modeling and algorithm of the average atlas construction are based on non-linear image registration and reconstruction. | |
| − | + | The algorithm does not depend on selecting a particular image as the template and the solution is optimal with respect to a minimization problem. | |
| + | In this project we want to construct the average atlas using a database with segmentations and validate the average atlas by the visual inspection of merged segmentations. | ||
| + | |||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | * We developed algorithms for the average atlas construction, finished programming in Matlab. | ||
| + | * We constructed the average atlas of 20 datasets for H&N and merged segmentations into the average atlas. | ||
| + | * We visualized the average atlas and its probability maps of segmentations in Slicer. | ||
</div> | </div> | ||
</div> | </div> | ||
==Project Results== | ==Project Results== | ||
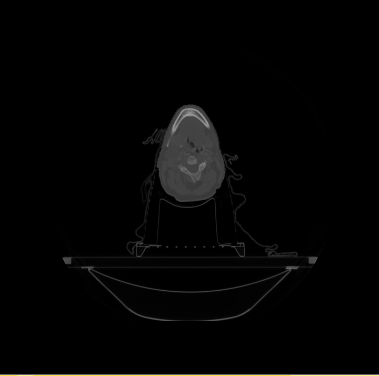
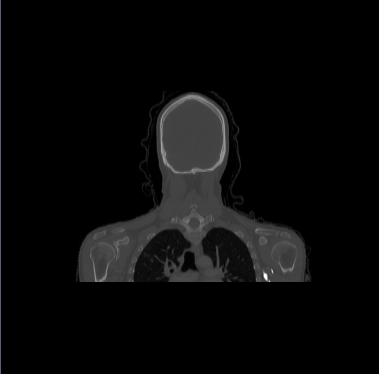
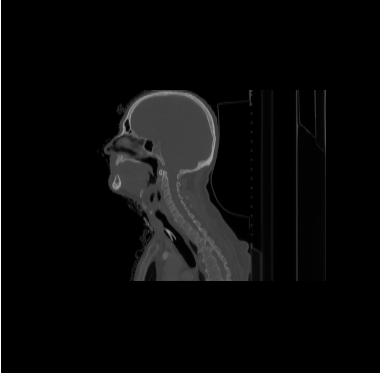
| − | * | + | * We have 20 datasets for H&N including lungs and chests. For example |
[[File:Axial_orig_image.png|||400px|||]] [[File:Coronal_orig_image.png|||400px|||]] [[File:Sagittal_orig_image.png|||400px|||]] | [[File:Axial_orig_image.png|||400px|||]] [[File:Coronal_orig_image.png|||400px|||]] [[File:Sagittal_orig_image.png|||400px|||]] | ||
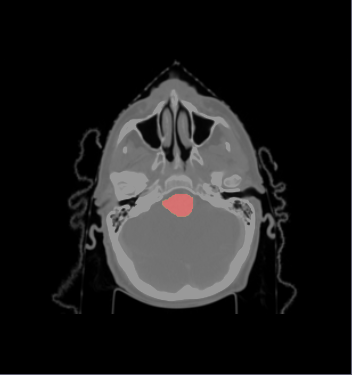
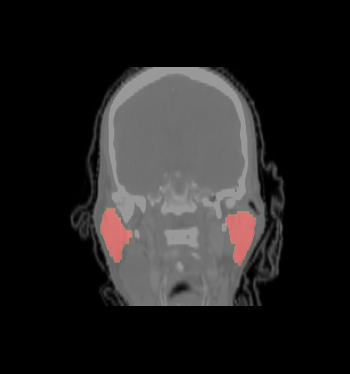
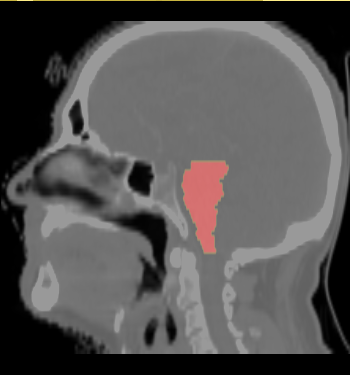
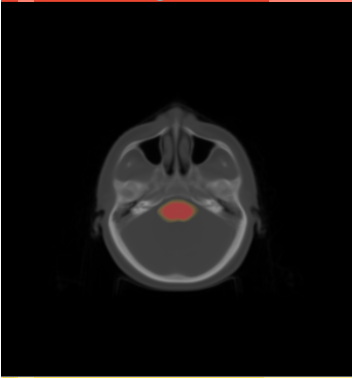
| − | * ROI of | + | * We selected the ROI of every data excluding lung and chest. For each data there exist the segmentations of brain stem, left and right parotids. For example |
[[File:Axial_roi_image.png|||400px|||]] [[File:Coronal_roi_image.png|||400px|||]] [[File:Sagittal_roi_image.png|||400px|||]] | [[File:Axial_roi_image.png|||400px|||]] [[File:Coronal_roi_image.png|||400px|||]] [[File:Sagittal_roi_image.png|||400px|||]] | ||
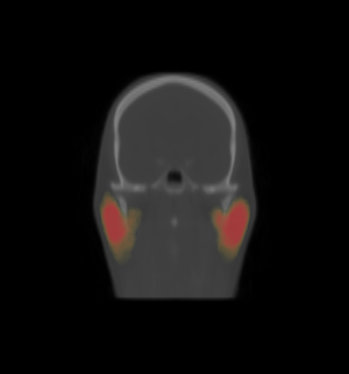
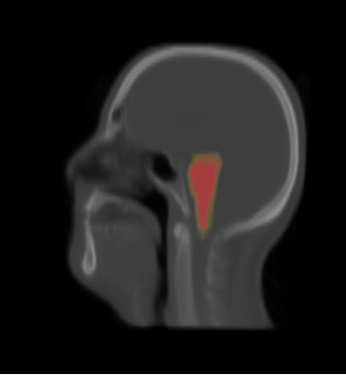
| − | * | + | * We constructed the average atlas of 20 datasets of ROI, and merged the segmentations into the average atlas. These segmentations are the probability maps of |
| + | brain stem, left and right parotids. | ||
| + | |||
[[File:Axial_atlas_image.png|||400px|||]] [[File:Coronal_atlas_image.png|||400px|||]] [[File:Sagittal_atlas_image.png|||400px|||]] | [[File:Axial_atlas_image.png|||400px|||]] [[File:Coronal_atlas_image.png|||400px|||]] [[File:Sagittal_atlas_image.png|||400px|||]] | ||
Latest revision as of 14:22, 27 June 2014
Home < 2014 Summer Project Week:Atlas SelectionKey Investigators
- Kanglin Chen (Fraunhofer MEVIS Germany)
- Gregory Sharp (MGH)
Project Description
Objective
Atlas selection is used for image segmentation. Normally, a single image is chosen as an atlas and the structures are segmented manually. The segmentation is transferred to patient data using non-linear image registration. However, image segmentation based on single image atlas is not stable. To improve the segmentation, we can select multiple images, which are suitable for image segmentation. After registration we can merge the segmentations to a final one. An idea to improve atlas selection process lies on that we take advantage of using an average atlas. The procedure of atlas selection using an average atlas for image segmentation is defined as follows:
- Construct an average atlas based on a database
- Every image of the database is aligned to the average atlas after the atlas construction.
- Register the average atlas to a new image and transfer the database and their segmentations to the new image.
- Compare the transformed database with the new image in the ROI and select some well-matched images with respected to some distance measure, e.g. MI, CC, NGF.
- Merge the segmentations of these well-matched images to a final one based on e.g. weighted voting or STAPLE algorithm.
The crucial step of atlas selection is the average atlas construction. The focus of this project lies on the construction of an average atlas using 3D datasets and validate the average atlas with the inspection of segmentations.
Approach, Plan
The modeling and algorithm of the average atlas construction are based on non-linear image registration and reconstruction. The algorithm does not depend on selecting a particular image as the template and the solution is optimal with respect to a minimization problem. In this project we want to construct the average atlas using a database with segmentations and validate the average atlas by the visual inspection of merged segmentations.
Progress
- We developed algorithms for the average atlas construction, finished programming in Matlab.
- We constructed the average atlas of 20 datasets for H&N and merged segmentations into the average atlas.
- We visualized the average atlas and its probability maps of segmentations in Slicer.
Project Results
- We have 20 datasets for H&N including lungs and chests. For example
- We selected the ROI of every data excluding lung and chest. For each data there exist the segmentations of brain stem, left and right parotids. For example
- We constructed the average atlas of 20 datasets of ROI, and merged the segmentations into the average atlas. These segmentations are the probability maps of
brain stem, left and right parotids.