Difference between revisions of "Projects:DiffeoMixedEffects"
Jfishbaugh (talk | contribs) |
Jfishbaugh (talk | contribs) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 8: | Line 8: | ||
Longitudinal image data has several sources of variability. First, there is inherent biological variability, both within a subject changing over time and also between subjects in a population. The goal of longitudinal analysis is to quantify this variability and make inferences about changes over time in a population. However, longitudinal imaging data also include unwanted sources of variability, such as noise in image acquisition, segmentation and registration errors, and human expert judgment, among others. These extraneous errors tend to dampen statistical power, especially when trying to distinguish between trajectories of two different populations, e.g., healthy and diseased. | Longitudinal image data has several sources of variability. First, there is inherent biological variability, both within a subject changing over time and also between subjects in a population. The goal of longitudinal analysis is to quantify this variability and make inferences about changes over time in a population. However, longitudinal imaging data also include unwanted sources of variability, such as noise in image acquisition, segmentation and registration errors, and human expert judgment, among others. These extraneous errors tend to dampen statistical power, especially when trying to distinguish between trajectories of two different populations, e.g., healthy and diseased. | ||
| − | [[File:Caudate_volume_plot.png|800px|center|thumb|'''Fig. | + | We study subcortical change associated with Huntington’s disease (HD), leveraging the longitudinal study PREDICT-HD. The longitudinal database consists of 65 female subjects: 23 controls (CTRL), 14 (LOW), 15 (MED), and 13 (HIGH). The LOW / MED / HIGH categories represent probability of onset of manifesting signs of HD. All subjects have had at least 3 MR images acquired approximately one year apart, with many subjects undergoing multiple scans per visit. Six subcortical pairs (caudate, putamen, hippocampus, thalamus, acumben, and pallidus) were segmented from each image and manually verified and cleaned. |
| + | |||
| + | The quality of each segmentation varies considerably for each time point, even when scans are obtained on the same day from the same scanner, as individual single-subject segmentation is prone to errors related to variability of imaging, image calibration, human expert judgment, and limited robustness of segmentation algorithms. While the segmentation quality is not easily assessed by viewing the 3D anatomical | ||
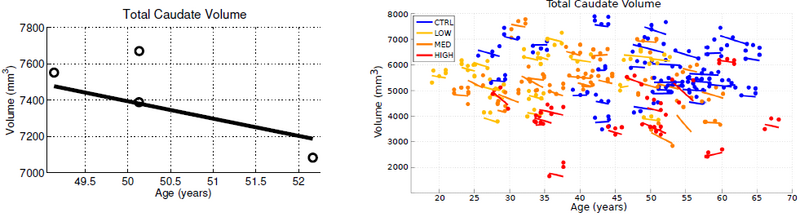
| + | surfaces, the temporal inconsistency becomes clear by investigating volume extracted from the shapes. The left side of Fig. 1 shows the variability in segmentation, illustrated by the temporal inconsistency of observed caudate volume, motivating the need for temporally consistent segmentations which properly account for correlated longitudinal data. | ||
| + | |||
| + | Continuous models of shape trajectory are estimated for each subject, resulting in personalized and temporally consistent anatomical evolution. Model estimation does not require point correspondence, facilitating the inclusion of all subcortical shapes simultaneously without imposing any topological constraints. Each subject’s personalized model allows us to generate shapes at any instant in time, from which desired shape properties, such as volume, can be extracted. We can therefore obtain a continuous evolution of volume for all subcortical structures without any explicit modeling of volume. Fig. 1 shows caudate volume extracted from each subject’s continuous shape model, demonstrating the flexibility of the shape model to capture both linear and non-linear volume trends with no prior assumption or constraint on linearity. Though | ||
| + | we only display caudate volume here, recall that each model is estimated by leveraging all shape data simultaneously (Fig. 2), which respects shape boundaries and locations, incorporating important geometric relationships between shapes. | ||
| + | |||
| + | [[File:Caudate_volume_plot.png|800px|center|thumb|'''Fig. 1.''' Left: For one subject, volume of observed caudates (open circles) and temporally consistent continuous volume extracted from diffeomorphic shape model (solid line). The difference in caudate volume extracted from scans obtained on the same day highlights the need for consistent segmentation. Right: Observed volume and volume extracted from diffeomorphic shape models for all 65 subjects. While the volume of the discrete shape observations show considerable variation, volume extracted from personalized models are continuous and temporally consistent.]] | ||
| + | |||
| + | '''Longitudinal analysis of striatal volume''' | ||
| + | |||
| + | We conduct a univariate analysis of volume extracted from shape, as striatal volume loss has been shown to be associated with the progression of HD. We aim to evaluate the benefit of spatiotemporal shape modeling, by comparing striatal volume extracted from the temporally | ||
| + | consistent shapes with volume extracted from the raw shape observations. Fig. 2 shows the results of linear mixed-effects analysis on striatal volumes for observed (left) and temporally consistent shapes (right), testing for the interaction between age and group membership. The estimated fixed-effects parameters for the temporally consistent (smoothed) category were found to be significant. This demonstrates the benefit of spatiotemporal shape modeling, as striatal volumes extracted from the temporally consistent shapes provide better separation between the control and LOW groups, and also between the control and HIGH groups. | ||
[[File:Observed_vs_consistent_striatal_volume.png|800px|center|thumb|'''Fig. 2.''' Longitudinal mixed-effects analysis of striatal volumes obtained from observed shapes (Left) and temporally consistent shapes (Right). Volume data are shown as filled black circles with corresponding individual trends. Note the improvement of the model fit in the consistent striatal volume over the observed striatal volume, which results in lower standard error of estimated fixed-effects parameters.]] | [[File:Observed_vs_consistent_striatal_volume.png|800px|center|thumb|'''Fig. 2.''' Longitudinal mixed-effects analysis of striatal volumes obtained from observed shapes (Left) and temporally consistent shapes (Right). Volume data are shown as filled black circles with corresponding individual trends. Note the improvement of the model fit in the consistent striatal volume over the observed striatal volume, which results in lower standard error of estimated fixed-effects parameters.]] | ||
| + | |||
| + | '''Longitudinal analysis of striatal shape''' | ||
| + | |||
| + | We perform a multivariate Hotelling’s T2 hypothesis test of the baseline shape (intercept) and trend (slope) between | ||
| + | controls and the combined HD groups.We compare the results for analyses using the original observed segmentations versus those obtained from spatiotemporal modeling as described in (Section 2.1). We represent these shapes in the particle optimization framework to estimate longitudinal fixed and random effects. Note that we do not normalize for size in these experiments, which means that we test for differences between control and combined HD groups based on both shape and size. | ||
| + | |||
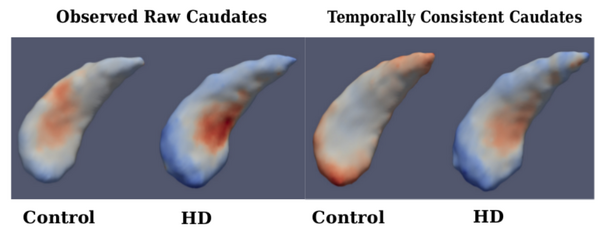
| + | Fig. 3 shows the estimated fixed-effects parameters for both groups, i.e., the baseline (intercept) shape with trajectory (slope) displayed as a color map. When comparing baseline shapes, we don’t find significant difference between controls and HD in either analysis. This is expected, as the onset of degeneration in HD is expected at a later age. But when comparing shape trends, we find significant differences between controls and HD for the temporally consistent shapes, but not in the case of raw shape observations. | ||
[[File:Compare_caudates.png|600px|center|thumb|'''Fig. 3.''' Left: Fixed-effects parameters for observed caudate shapes (Far Left-Control, Mid Left- HD), Right: Fixed-effects parameters for temporally consistent caudate shapes (Mid Right-Control, Far Right-HD); Fixed effects slope: Blue-Red indicates Local Contraction - Expansion.]] | [[File:Compare_caudates.png|600px|center|thumb|'''Fig. 3.''' Left: Fixed-effects parameters for observed caudate shapes (Far Left-Control, Mid Left- HD), Right: Fixed-effects parameters for temporally consistent caudate shapes (Mid Right-Control, Far Right-HD); Fixed effects slope: Blue-Red indicates Local Contraction - Expansion.]] | ||
Latest revision as of 05:56, 14 October 2014
Home < Projects:DiffeoMixedEffectsBack to Utah 2 Algorithms
Ongoing Work
Diffeomorphic Shape Trajectories for Improved Longitudinal Segmentation and Statistics
Longitudinal image data has several sources of variability. First, there is inherent biological variability, both within a subject changing over time and also between subjects in a population. The goal of longitudinal analysis is to quantify this variability and make inferences about changes over time in a population. However, longitudinal imaging data also include unwanted sources of variability, such as noise in image acquisition, segmentation and registration errors, and human expert judgment, among others. These extraneous errors tend to dampen statistical power, especially when trying to distinguish between trajectories of two different populations, e.g., healthy and diseased.
We study subcortical change associated with Huntington’s disease (HD), leveraging the longitudinal study PREDICT-HD. The longitudinal database consists of 65 female subjects: 23 controls (CTRL), 14 (LOW), 15 (MED), and 13 (HIGH). The LOW / MED / HIGH categories represent probability of onset of manifesting signs of HD. All subjects have had at least 3 MR images acquired approximately one year apart, with many subjects undergoing multiple scans per visit. Six subcortical pairs (caudate, putamen, hippocampus, thalamus, acumben, and pallidus) were segmented from each image and manually verified and cleaned.
The quality of each segmentation varies considerably for each time point, even when scans are obtained on the same day from the same scanner, as individual single-subject segmentation is prone to errors related to variability of imaging, image calibration, human expert judgment, and limited robustness of segmentation algorithms. While the segmentation quality is not easily assessed by viewing the 3D anatomical surfaces, the temporal inconsistency becomes clear by investigating volume extracted from the shapes. The left side of Fig. 1 shows the variability in segmentation, illustrated by the temporal inconsistency of observed caudate volume, motivating the need for temporally consistent segmentations which properly account for correlated longitudinal data.
Continuous models of shape trajectory are estimated for each subject, resulting in personalized and temporally consistent anatomical evolution. Model estimation does not require point correspondence, facilitating the inclusion of all subcortical shapes simultaneously without imposing any topological constraints. Each subject’s personalized model allows us to generate shapes at any instant in time, from which desired shape properties, such as volume, can be extracted. We can therefore obtain a continuous evolution of volume for all subcortical structures without any explicit modeling of volume. Fig. 1 shows caudate volume extracted from each subject’s continuous shape model, demonstrating the flexibility of the shape model to capture both linear and non-linear volume trends with no prior assumption or constraint on linearity. Though we only display caudate volume here, recall that each model is estimated by leveraging all shape data simultaneously (Fig. 2), which respects shape boundaries and locations, incorporating important geometric relationships between shapes.
Longitudinal analysis of striatal volume
We conduct a univariate analysis of volume extracted from shape, as striatal volume loss has been shown to be associated with the progression of HD. We aim to evaluate the benefit of spatiotemporal shape modeling, by comparing striatal volume extracted from the temporally consistent shapes with volume extracted from the raw shape observations. Fig. 2 shows the results of linear mixed-effects analysis on striatal volumes for observed (left) and temporally consistent shapes (right), testing for the interaction between age and group membership. The estimated fixed-effects parameters for the temporally consistent (smoothed) category were found to be significant. This demonstrates the benefit of spatiotemporal shape modeling, as striatal volumes extracted from the temporally consistent shapes provide better separation between the control and LOW groups, and also between the control and HIGH groups.

Longitudinal analysis of striatal shape
We perform a multivariate Hotelling’s T2 hypothesis test of the baseline shape (intercept) and trend (slope) between controls and the combined HD groups.We compare the results for analyses using the original observed segmentations versus those obtained from spatiotemporal modeling as described in (Section 2.1). We represent these shapes in the particle optimization framework to estimate longitudinal fixed and random effects. Note that we do not normalize for size in these experiments, which means that we test for differences between control and combined HD groups based on both shape and size.
Fig. 3 shows the estimated fixed-effects parameters for both groups, i.e., the baseline (intercept) shape with trajectory (slope) displayed as a color map. When comparing baseline shapes, we don’t find significant difference between controls and HD in either analysis. This is expected, as the onset of degeneration in HD is expected at a later age. But when comparing shape trends, we find significant differences between controls and HD for the temporally consistent shapes, but not in the case of raw shape observations.
Literature
[1] Muralidharan, P., Fishbaugh, J., Johnson, H., Durrleman, S., Paulsen, J., Gerig, G., Fletcher, P.T. Diffeomorphic shape trajectories for improved longitudinal segmentation and statistics. Proc. of Medical Image Computing and Computer Assisted Intervention (MICCAI '14). (2014).
Key Investigators
- Utah: Prasanna Muralidharan, James Fishbaugh, P.T. Fletcher, Guido Gerig
- INRIA/ICM, Pitie Salpetriere Hospital: Stanley Durrleman
- Department of Psychiatry, Carver College of Medicine, University of Iowa: Jane Paulsen, Hans Johnson