Difference between revisions of "CIP and Nipype"
From NAMIC Wiki
| (2 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
==Key Investigators== | ==Key Investigators== | ||
| − | + | Rola Harmouche, James Ross, Alex Yarmakovich | |
==Project Description== | ==Project Description== | ||
| Line 18: | Line 18: | ||
* Define and implement a set of workflows (nipype/Vistrails) for the following tasks: | * Define and implement a set of workflows (nipype/Vistrails) for the following tasks: | ||
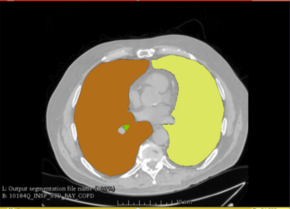
** computing body composition (ex pectoralis muscles, subcutaneous fat, visceral fat, paravertebral muscles...) phenotypes from pre-labeled CT data. The phenotypes consist of cross sectional areas of each label and CT intensity statistics within the labeled region | ** computing body composition (ex pectoralis muscles, subcutaneous fat, visceral fat, paravertebral muscles...) phenotypes from pre-labeled CT data. The phenotypes consist of cross sectional areas of each label and CT intensity statistics within the labeled region | ||
| − | [[File: | + | [[File:LungParenchyma.png|290px]] |
** computing lung parenchyma phenotypes from CT data | ** computing lung parenchyma phenotypes from CT data | ||
| Line 26: | Line 26: | ||
* Generated nipype interfaces for the CLIs available as part of the chest imaging platform and for python and for python classes | * Generated nipype interfaces for the CLIs available as part of the chest imaging platform and for python and for python classes | ||
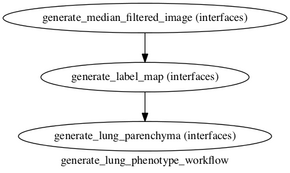
* Implemented an example workflow for the generation of lung parenchyma phenotypes | * Implemented an example workflow for the generation of lung parenchyma phenotypes | ||
| − | [[File: | + | [[File:parenchyma_workflow_graph.dot.png|290px]] |
| + | [[File:command_line.png|290px]] | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 16:27, 9 January 2015
Home < CIP and NipypeKey Investigators
Rola Harmouche, James Ross, Alex Yarmakovich
Project Description
Objective
- We now have a suite of CLIs and python scripts for the processing and the analysis of chest images ready to be incorporated in Slicer as part of the Chest Imaging Platform Extension. This week we will be specifically focusing on defining clinically relevant chest image processing workflows that utilize the CLIs and scripts and implementing the workflows in nipype or Vistrails for their deployment in high performance computing environments.
Approach, Plan
- Our first task is to generate nipype interfaces from the slicer CLIs and python scripts
- Define and implement a set of workflows (nipype/Vistrails) for the following tasks:
- computing body composition (ex pectoralis muscles, subcutaneous fat, visceral fat, paravertebral muscles...) phenotypes from pre-labeled CT data. The phenotypes consist of cross sectional areas of each label and CT intensity statistics within the labeled region
- computing lung parenchyma phenotypes from CT data