Difference between revisions of "2015 Summer Project Week:LungCAD"
From NAMIC Wiki
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (6 intermediate revisions by one other user not shown) | |||
| Line 10: | Line 10: | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * Jayender Jagadeesan | + | * Jayender Jagadeesan (BWH) |
| − | * Tobias Penskofer | + | * Tobias Penskofer (Charite, Berlin) |
| − | * Sandy Wells | + | * Sandy Wells (BWH) |
| − | * Clara | + | * Clara Meiner (previously BWH) |
| + | * Raul San Jose Estepar (BWH) | ||
| + | * Jorge Onieva (BWH) | ||
==Project Description== | ==Project Description== | ||
| Line 20: | Line 22: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
* Develop a module in 3D Slicer to segment the ground glass opacity (GGO) tumor, apply [https://www.slicer.org/slicerWiki/index.php/Documentation/Nightly/Modules/HeterogeneityCAD HeterogeneityCAD] to obtain imaging metrics and classify the GGO. | * Develop a module in 3D Slicer to segment the ground glass opacity (GGO) tumor, apply [https://www.slicer.org/slicerWiki/index.php/Documentation/Nightly/Modules/HeterogeneityCAD HeterogeneityCAD] to obtain imaging metrics and classify the GGO. | ||
| − | * Provide the module as an extension part of [ | + | * Provide the module as an extension part of [https://www.slicer.org/wiki/Documentation/Nightly/Extensions/OpenCAD OpenCAD] |
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
| Line 31: | Line 33: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
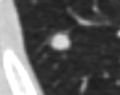
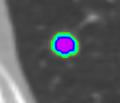
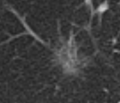
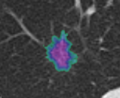
* Completed analysis for 248 GGOs and trained SVM with an accuracy of 89% to classify GGOs | * Completed analysis for 248 GGOs and trained SVM with an accuracy of 89% to classify GGOs | ||
| + | * Developed the framework for the LungCAD module in Slicer | ||
| + | * Evaluated the Lesion segmentation algorithm as part of the Chest Imaging Platform | ||
| + | * Segmentation works well and is able to prevent the segmentation of vessels running through the lesion | ||
| + | * LungCAD calls Lesion Segmentation CLI for segmentation and HeterogeneityCAD to evaluate features | ||
| + | * Pre-processed SVM will be utilized to classify the GGOs | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 18:07, 10 July 2017
Home < 2015 Summer Project Week:LungCADKey Investigators
- Jayender Jagadeesan (BWH)
- Tobias Penskofer (Charite, Berlin)
- Sandy Wells (BWH)
- Clara Meiner (previously BWH)
- Raul San Jose Estepar (BWH)
- Jorge Onieva (BWH)
Project Description
Objective
- Develop a module in 3D Slicer to segment the ground glass opacity (GGO) tumor, apply HeterogeneityCAD to obtain imaging metrics and classify the GGO.
- Provide the module as an extension part of OpenCAD
Approach, Plan
- Implement a simple region growing algorithm
- Apply HeterogeneityCAD module
- Use predetermined SVM classifier to decide the lesion type
Progress
- Completed analysis for 248 GGOs and trained SVM with an accuracy of 89% to classify GGOs
- Developed the framework for the LungCAD module in Slicer
- Evaluated the Lesion segmentation algorithm as part of the Chest Imaging Platform
- Segmentation works well and is able to prevent the segmentation of vessels running through the lesion
- LungCAD calls Lesion Segmentation CLI for segmentation and HeterogeneityCAD to evaluate features
- Pre-processed SVM will be utilized to classify the GGOs