Difference between revisions of "2017 Winter Project Week/Needle Segmentation from MRI"
From NAMIC Wiki
(first draft) |
|||
| (9 intermediate revisions by 4 users not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-Winter2017.png|[[2017_Winter_Project_Week#Projects|Projects List]] | Image:PW-Winter2017.png|[[2017_Winter_Project_Week#Projects|Projects List]] | ||
| + | Image:needles_winter_project_week_2017.png | ||
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
* Ziyang Wang, BWH | * Ziyang Wang, BWH | ||
| − | * Alireza Mehrtash | + | * Alireza Mehrtash, BWH |
| − | * Alireza Ziaei Torbati | + | * Alireza Ziaei Torbati, BWH |
* Guillaume Pernelle, Imperial College London | * Guillaume Pernelle, Imperial College London | ||
| + | * Paolo Zaffino, Magna Graecia University of Catanzaro, Italy | ||
| + | * Andre Mastmeyer, University of Luebeck, Germany | ||
* Tina Kapur, BWH/HMS | * Tina Kapur, BWH/HMS | ||
| + | |||
| + | ==Deep Learning Demo Result== | ||
| + | [[Media:Needle video.avi]] | ||
==Project Description== | ==Project Description== | ||
| − | This project is a continuation of the project started during the 2016 summer project week ([[2016 Summer Project Week/Needle Segmentation from MRI | Needle Segmentation from MRI]]), | + | This project is a continuation of the project started during the 2016 summer project week ([[2016 Summer Project Week/Needle Segmentation from MRI | Needle Segmentation from MRI]]), [http://needlefinder.org NeedleFinder] offers tools to segment needles from MRI/CT. It has mostly been tested on MRI from GYN brachytherapy cases. Currently the user must provide the needle tip for the segmentation to start. We aim to detect the tip automatically to make the needle segmentation fully automatic. |
| − | NeedleFinder offers tools to segment needles from MRI/CT. It has mostly been tested on MRI from GYN brachytherapy cases. Currently the user must provide the needle tip for the segmentation to start. We aim to detect the tip automatically to make the needle segmentation fully automatic. | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 25: | Line 30: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
* We have manually segmented around 1k needles from GYN brachytherapy cases. We want to use this data in a supervised learning approach. | * We have manually segmented around 1k needles from GYN brachytherapy cases. We want to use this data in a supervised learning approach. | ||
| − | * We | + | * We have a pipeline for preprocessing the data (image spacing and normalization) |
| + | * TODO: choose between 2 strategies: | ||
| + | ** Continue with the binary classification approach -> in the region of interest, voxels are classified as positive (needle) or negative (background). Challenge: link those voxels together to form needles / filter out false negatives. Alternative: try to find the tip position and use the previous algorithm to segment the needle | ||
| + | ** Use a semantic segmentation approach (fully convolutional neural networks [J Long 2015], Region-based semantic segmentation with end-to-end training [Caesar 2016]) | ||
| + | |||
... | ... | ||
| Line 31: | Line 40: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
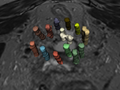
| − | + | * Many useful discussions | |
| + | * The workflow was modified in order to segment the entire needle instead of just the tip | ||
| + | * Good preliminary results (see picture and [[Media:Needle video.avi]]) | ||
| + | * Start to increase the patch size and the number of negative patches | ||
| + | * Start to test U-net strategy | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 22:53, 12 January 2017
Home < 2017 Winter Project Week < Needle Segmentation from MRIKey Investigators
- Ziyang Wang, BWH
- Alireza Mehrtash, BWH
- Alireza Ziaei Torbati, BWH
- Guillaume Pernelle, Imperial College London
- Paolo Zaffino, Magna Graecia University of Catanzaro, Italy
- Andre Mastmeyer, University of Luebeck, Germany
- Tina Kapur, BWH/HMS
Deep Learning Demo Result
Project Description
This project is a continuation of the project started during the 2016 summer project week ( Needle Segmentation from MRI), NeedleFinder offers tools to segment needles from MRI/CT. It has mostly been tested on MRI from GYN brachytherapy cases. Currently the user must provide the needle tip for the segmentation to start. We aim to detect the tip automatically to make the needle segmentation fully automatic.
Objective
- Automatic detection of needle tips in MRI from GYN brachytherapy cases.
Approach, Plan
- We have manually segmented around 1k needles from GYN brachytherapy cases. We want to use this data in a supervised learning approach.
- We have a pipeline for preprocessing the data (image spacing and normalization)
- TODO: choose between 2 strategies:
- Continue with the binary classification approach -> in the region of interest, voxels are classified as positive (needle) or negative (background). Challenge: link those voxels together to form needles / filter out false negatives. Alternative: try to find the tip position and use the previous algorithm to segment the needle
- Use a semantic segmentation approach (fully convolutional neural networks [J Long 2015], Region-based semantic segmentation with end-to-end training [Caesar 2016])
...
Progress
- Many useful discussions
- The workflow was modified in order to segment the entire needle instead of just the tip
- Good preliminary results (see picture and Media:Needle video.avi)
- Start to increase the patch size and the number of negative patches
- Start to test U-net strategy