Difference between revisions of "Project Week 25/Multimodal:"
From NAMIC Wiki
Jfishbaugh (talk | contribs) |
|||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 8: | Line 8: | ||
==Project Description== | ==Project Description== | ||
| − | Project details can downloaded here as [http://research.engineering.nyu.edu/~fishbaugh/docs/Gerig-Slicer-6-26-2017-final.pptx pptx] or [ | + | * '''Project details can downloaded here as [http://research.engineering.nyu.edu/~fishbaugh/docs/Gerig-Slicer-6-26-2017-final.pptx pptx] or [https://na-mic.org/w/images/a/ae/Gerig-Slicer-6-26-2017-final.pdf pdf].''' |
| + | * '''Download a [https://na-mic.org/w/images/a/af/Tutorial_HOA.pdf Slicer tutorial].''' | ||
| + | |||
{| class="wikitable" | {| class="wikitable" | ||
| Line 18: | Line 20: | ||
<!-- Objective bullet points --> | <!-- Objective bullet points --> | ||
| − | 3D/4D Ophthalmology Image Anaylsis Framework | + | 3D/4D Ophthalmology Image Anaylsis Framework: Continuation of initial January 2017 project by Sungmin Hong: |
* Read 4D hyperspectral data | * Read 4D hyperspectral data | ||
* Viewer and interactor for 3D hi-res image data and 4D hyperspectral data | * Viewer and interactor for 3D hi-res image data and 4D hyperspectral data | ||
| Line 36: | Line 38: | ||
** Implement/integrate 4D hyperspectral data viewer to show image and spectral information. | ** Implement/integrate 4D hyperspectral data viewer to show image and spectral information. | ||
** Integrate registration functionality for co-registration between 3D hi-res data and 4D hyperspectral data at image level | ** Integrate registration functionality for co-registration between 3D hi-res data and 4D hyperspectral data at image level | ||
| − | ** | + | ** Explore most recent "Segment Editor" features for cell and organelle segmentation |
| − | |||
* User Manual | * User Manual | ||
** Create an user manual to comprehend a overview of an extension | ** Create an user manual to comprehend a overview of an extension | ||
| − | |||
| | | | ||
<!-- Progress and Next steps bullet points (fill out at the end of project week --> | <!-- Progress and Next steps bullet points (fill out at the end of project week --> | ||
| − | * | + | * Manual |
| − | ** | + | ** [https://na-mic.org/w/images/a/af/Tutorial_HOA.pdf Slicer tutorial] explaining multi-volume conversion and 3D/4D registration. |
| − | |||
| − | |||
* Registration | * Registration | ||
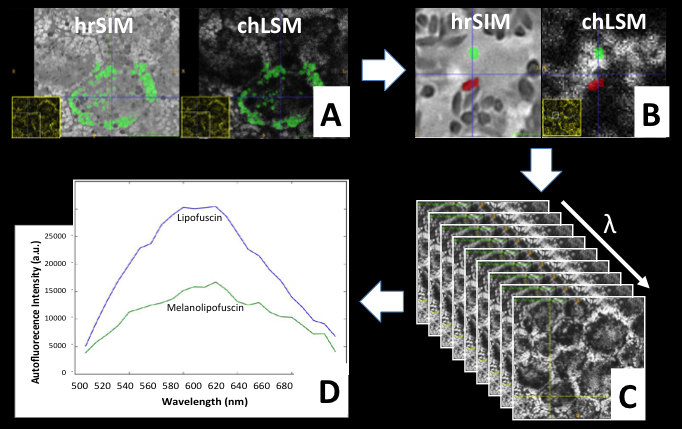
| − | ** Basic registration algorithms in Slicer worked well | + | ** '''Basic registration algorithms in Slicer worked well for linear registration''' between 3D SIM and cropped and dimension reduced 4D LSM data. |
| − | ** | + | ** Automatic detection of corresponding subregions between hires 3D SIM and low-res 4D LSM data may be implemented via autocorrelation. |
* Segmentation | * Segmentation | ||
| − | ** | + | ** '''GrowCut segmentation worked well for 3D segmentation''' of organelles of different contrast, e.g. dark Lipofuscin and bright Melanolipofuscin as required by clinical researchers. |
| − | ** | + | ** Cell segmentation/outlining and combination with organelle segmentation can be solved with the Segment Editor features. |
| − | ** | + | |
| + | * Hyperspectral Analysis | ||
| + | ** Module to convert 4D LSM data to a series of 3D data compatible to MultiVolume Explorer was implemented at Jan. 2017 project week. | ||
| + | ** With such series of 3D data, MultiVolumeExplorer offers very basic qualitative analysis hyperspectral data by plotting a spectral curve per voxel. | ||
| + | ** '''Quantitative label statistics''' by combining 3D segmentation labels with 4D hyperspectral data '''so far solved via external python script''', will need to be added to the multivolume explorer in the future. | ||
| + | |||
|} | |} | ||
| Line 66: | Line 69: | ||
==Background and References== | ==Background and References== | ||
<!-- Use this space for information that may help people better understand your project, like links to papers, source code, or data --> | <!-- Use this space for information that may help people better understand your project, like links to papers, source code, or data --> | ||
| + | |||
| + | [1] Thomas Ach, Sungmin Hong, Rainer Heintzmann, Jost Hillenkamp, Kenneth R. Sloan, Neel Dey, Guido Gerig, R. Theodore Smith, Christine A. Curcio, Katharina Bermond, High-resolution and hyperspectral imaging of autofluorescent retinal pigment epithelium (RPE) granules, ARVO 2017 Annual Meeting Abstracts, No. 3382, [http://www.arvo.org/webs/am2017/sectionpdf/RC/Session%20363%20Macular%20degeneration-cell%20biology.pdf Abstract] | ||
| + | |||
| + | [2] Tong Y, Ben Ami T, Hong S, Heintzmann R, Gerig G, Ablonczy Z, Curcio CA, Ach T, Smith RT., HYPERSPECTRAL AUTOFLUORESCENCE IMAGING OF DRUSEN AND RETINAL PIGMENT EPITHELIUM IN DONOR EYES WITH AGE-RELATED MACULAR DEGENERATION, Retina. 2016 Dec;36 Suppl 1:S127-S136, [http://journals.lww.com/retinajournal/Citation/2016/12001/HYPERSPECTRAL_AUTOFLUORESCENCE_IMAGING_OF_DRUSEN.14.aspx Paper] | ||
Latest revision as of 10:29, 30 June 2017
Home < Project Week 25 < Multimodal:Back to Projects List
Key Investigators
- Guido Gerig (NYU Tandon School of Engineering, USA)
- Sungmin Hong (NYU Tandon School of Engineering, USA)
Project Description
- Project details can downloaded here as pptx or pdf.
- Download a Slicer tutorial.
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
3D/4D Ophthalmology Image Anaylsis Framework: Continuation of initial January 2017 project by Sungmin Hong:
|
|
|
Illustrations
Background and References
[1] Thomas Ach, Sungmin Hong, Rainer Heintzmann, Jost Hillenkamp, Kenneth R. Sloan, Neel Dey, Guido Gerig, R. Theodore Smith, Christine A. Curcio, Katharina Bermond, High-resolution and hyperspectral imaging of autofluorescent retinal pigment epithelium (RPE) granules, ARVO 2017 Annual Meeting Abstracts, No. 3382, Abstract
[2] Tong Y, Ben Ami T, Hong S, Heintzmann R, Gerig G, Ablonczy Z, Curcio CA, Ach T, Smith RT., HYPERSPECTRAL AUTOFLUORESCENCE IMAGING OF DRUSEN AND RETINAL PIGMENT EPITHELIUM IN DONOR EYES WITH AGE-RELATED MACULAR DEGENERATION, Retina. 2016 Dec;36 Suppl 1:S127-S136, Paper