Difference between revisions of "Algorithm:MIT:New"
| Line 21: | Line 21: | ||
| | [[Image:brain.png|thumb|left|200px]] | | | [[Image:brain.png|thumb|left|200px]] | ||
| | | | | | ||
| + | |||
| + | == [[Algorithm:MIT:Spherical Wavelets |Spherical Wavelets]] == | ||
| + | |||
| + | This type of algorithms assigns a tissue type to each voxel in the volume. Incorporating prior shape information biases the label assignment towards contiguous regions that are consistent with the shape model. [[Algorithm:MIT:Shape_Based_Segmentation_And_Registration|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> K.M. Pohl, J. Fisher, S. Bouix, M. Shenton, R. W. McCarley, W.E.L. Grimson, R. Kikinis, and W.M. Wells. Using the Logarithm of Odds to Define a Vector Space on Probabilistic Atlases. Accepted to the Special Issue of Best Selected Papers from MICCAI 06 in Medical Image Analysis [[Algorithm:MIT:Shape_Based_Segmentation_And_Registration#Publications|More...]] | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:brain.png|thumb|left|200px]] | ||
| + | | | | ||
== [[Algorithm:MIT:Shape_Based_Level_Set_Segmentation|Shape Based Level Segmentation]] == | == [[Algorithm:MIT:Shape_Based_Level_Set_Segmentation|Shape Based Level Segmentation]] == | ||
Revision as of 18:39, 9 November 2007
Home < Algorithm:MIT:NewBack to NA-MIC Algorithms
Contents
- 1 Overview of MIT Algorithms
- 2 MIT Projects
- 2.1 Shape Based Segmentation and Registration
- 2.2 Spherical Wavelets
- 2.3 Shape Based Level Segmentation
- 2.4 Registration Regularization
- 2.5 DTI Fiber Clustering and Fiber-Based Analysis
- 2.6 Fiber Tract Modeling, Clustering, and Quantitative Analysis
- 2.7 DTI-based Segmentation
- 2.8 Fiber-Tract-Bundle-based Non-Linear Registration
- 2.9 Stochastic Tractography
- 2.10 fMRI Detection and Analysis
- 2.11 Population Analysis of Anatomical Variability
- 2.12 Groupwise Registration
Overview of MIT Algorithms
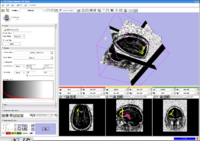
A brief overview of the MIT's algorithms goes here. This should not be much longer than a paragraph. Remember that people visiting your site want to be able to understand very quickly what you're all about and then they want to jump into your site's projects. The projects below are organized into a two column table: the left column is for representative images and the right column is for project overviews. The number of rows corresponds to the number of projects at your site. Put the most interesting and relevant projects at the top of the table. You do not need to organize the table according to subject matter (i.e. do not group all segmentation projects together and all DWI projects together).
MIT Projects
Shape Based Segmentation and RegistrationThis type of algorithms assigns a tissue type to each voxel in the volume. Incorporating prior shape information biases the label assignment towards contiguous regions that are consistent with the shape model. More... New: K.M. Pohl, J. Fisher, S. Bouix, M. Shenton, R. W. McCarley, W.E.L. Grimson, R. Kikinis, and W.M. Wells. Using the Logarithm of Odds to Define a Vector Space on Probabilistic Atlases. Accepted to the Special Issue of Best Selected Papers from MICCAI 06 in Medical Image Analysis More... | |
Spherical WaveletsThis type of algorithms assigns a tissue type to each voxel in the volume. Incorporating prior shape information biases the label assignment towards contiguous regions that are consistent with the shape model. More... New: K.M. Pohl, J. Fisher, S. Bouix, M. Shenton, R. W. McCarley, W.E.L. Grimson, R. Kikinis, and W.M. Wells. Using the Logarithm of Odds to Define a Vector Space on Probabilistic Atlases. Accepted to the Special Issue of Best Selected Papers from MICCAI 06 in Medical Image Analysis More... | |
Shape Based Level SegmentationThis class of algorithms explicitly manipulates the representation of the object boundary to fit the strong gradients in the image, indicative of the object outline. Bias in the boundary evolution towards the likely shapes improves the robustness of the segmentation results when the intensity information alone is insufficient for boundary detection. More... New: | |
Registration RegularizationWe are interested in the effects of registration regularization on segmentation accuracy in joint registration-segmentation. More... New: Submission for MICCAI 2007 | |
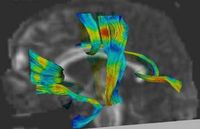
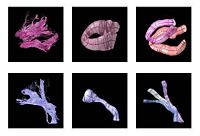
DTI Fiber Clustering and Fiber-Based AnalysisThe goal of this project is to provide structural description of the white matter architecture as a partition into coherent fiber bundles and clusters, and to use these bundles for quantitative measurement. More... New: Monica E. Lemmond, Lauren J. O'Donnell, Stephen Whalen, Alexandra J. Golby. Characterizing Diffusion Along White Matter Tracts Affected by Primary Brain Tumors. Accepted to HBM 2007. | |
Fiber Tract Modeling, Clustering, and Quantitative AnalysisThe goal of this work is to model the shape of the fiber bundles and use this model discription in clustering and statistical analysis of fiber tracts. More... New: M. Maddah, W. M. Wells, S. K. Warfield, C.-F. Westin, and W. E. L. Grimson, Probabilistic Clustering and Quantitative Analysis of White Matter Fiber Tracts,IPMI 2007, Netherlands. | |
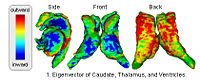
DTI-based SegmentationUnlike conventional MRI, DTI provides adequate contrast to segment the thalamic nuclei, which are gray matter structures. More... New: Ulas Ziyan, David Tuch, Carl-Fredrik Westin. Segmentation of Thalamic Nuclei from DTI using Spectral Clustering. Accepted to MICCAI 2006. | |
Fiber-Tract-Bundle-based Non-Linear RegistrationThe goal of this work is to utilize the anatomical information from segmented fiber bundles and use this information for registering fiber tracts and the underlying DTI images. More... New: | |
Stochastic TractographyThis work calculates posterior distributions of white matter fiber tract parameters given diffusion observations in a DWI volume. More... New: | |
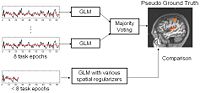
fMRI Detection and AnalysisWe are exploring algorithms for improved fMRI detection and interpretation by incorporting spatial priors and anatomical information to guide the detection. More... New: Wanmei Ou, Sandy Wells, Polina Golland. Bridging Spatial Regularization And Anatomical Priors in fMRI Detection. In preparation for submission to IEEE TMI. | |
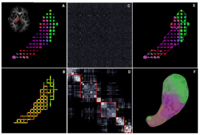
Population Analysis of Anatomical VariabilityOur goal is to develop mathematical approaches to modeling anatomical variability within and across populations using tools like local shape descriptors of specific regions of interest and global constellation descriptors of multiple ROI's. More... New: Mert R Sabuncu and Polina Golland. Structural Constellations for Population Analysis of Anatomical Variability. | |
Groupwise RegistrationWe are exploring algorithms for groupwise registration of medical data. More... New: Serdar K Balci, Polina Golland, Sandy Wells, Lilla Zollei, Mert R Sabuncu and Kinh Tieu. Groupwise registration of medical data. |