Difference between revisions of "DBP2:MIND:itkBayesianLesion"
From NAMIC Wiki
Hjbockholt (talk | contribs) |
Hjbockholt (talk | contribs) |
||
| Line 3: | Line 3: | ||
* stage one uses k-means tissue classification into grey, white, or csf of co-registered T1/T2 | * stage one uses k-means tissue classification into grey, white, or csf of co-registered T1/T2 | ||
* stage two use bayesian classifier into normal tissue or lesion using results of stage one and co-reigstered Flair | * stage two use bayesian classifier into normal tissue or lesion using results of stage one and co-reigstered Flair | ||
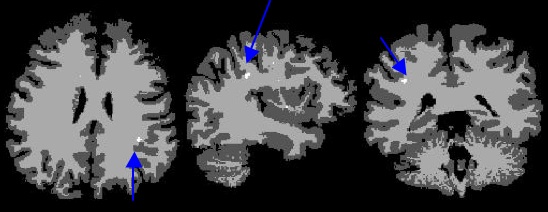
| + | Below image shows current result using T1,T2, and FLAIR, segmented into gray, white, csf, and lesion, the blue arrow highlights correct location of lesion | ||
| + | |||
[[Image:ItkBayesianLesion_example_results.jpg]] | [[Image:ItkBayesianLesion_example_results.jpg]] | ||
Latest revision as of 13:27, 18 March 2008
Home < DBP2:MIND:itkBayesianLesionITK-based 2 stage lesion segmentation method developed by Vince Magnotta
* stage one uses k-means tissue classification into grey, white, or csf of co-registered T1/T2 * stage two use bayesian classifier into normal tissue or lesion using results of stage one and co-reigstered Flair
Below image shows current result using T1,T2, and FLAIR, segmented into gray, white, csf, and lesion, the blue arrow highlights correct location of lesion