Difference between revisions of "NA-MIC Brains Collaboration"
From NAMIC Wiki
| Line 30: | Line 30: | ||
Here is a list of tools that are being developed as part of this project. A number of these tools have been contributed to the NITRC website. All of the tools are built upon the Slicer3 execution model and therefore will readily plug-in to Slicer3. | Here is a list of tools that are being developed as part of this project. A number of these tools have been contributed to the NITRC website. All of the tools are built upon the Slicer3 execution model and therefore will readily plug-in to Slicer3. | ||
*BRAINSFit - Linear fitting of two 3D image sets using a mutual information similarity metric | *BRAINSFit - Linear fitting of two 3D image sets using a mutual information similarity metric | ||
| − | *BRAINSDemonsWarp - Non-linear registration of two image sets. This includes the diffeomorphic demons algorithm along with a number of useful enhancements. | + | *[http://www.nitrc.org/projects/brainsdemonwarp/ BRAINSDemonsWarp] - Non-linear registration of two image sets. This includes the diffeomorphic demons algorithm along with a number of useful enhancements. |
*[http://www.nitrc.org/projects/brainsmush/ BRAINSMush] - Tool to combine T1 and T2 datasets into a composite image. The resulting image is then used to perform brain extraction. | *[http://www.nitrc.org/projects/brainsmush/ BRAINSMush] - Tool to combine T1 and T2 datasets into a composite image. The resulting image is then used to perform brain extraction. | ||
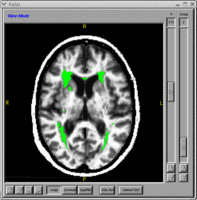
*Neural Networks - Automated segmentation of subcortical and cerebellar regions of interest using an artificial neural network. | *Neural Networks - Automated segmentation of subcortical and cerebellar regions of interest using an artificial neural network. | ||
Revision as of 22:12, 9 December 2008
Home < NA-MIC Brains CollaborationBack to NA-MIC_External_Collaborations
Contents
Abstract
This project is not an NCBC collaboration grant, but instead a Continued Development and Maintenance of Software grant. The intent of this application is to update the BRAINS image analysis software developed at the University of Iowa.
Grant #
1R01NS050568
Key Personnel
The collaborators include Vincent Magnotta, Hans Johnson, Jeremy Bockholt, and Nancy Andreasen.
Projects
There are three main thrusts of this application as outlined below.
Implement an Automated Brain Analysis Pipeline
Code Refactoring for Cross Platform Support
- Refactor Current Modules using ITK and VTK
- Build GUI using Slicer3 Interface
- Use Python as Scripting Language
Validation of Pipeline Using MIND MCIC Sample
- Evaluate MIND MCIC Human Phantom Sample Using Automated Pipeline
- Evaluate Differences between Patients with Schizophrenia and Normal Controls using Automated Pipeline
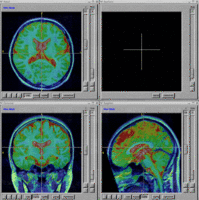
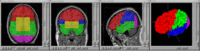
Tools Developed
Here is a list of tools that are being developed as part of this project. A number of these tools have been contributed to the NITRC website. All of the tools are built upon the Slicer3 execution model and therefore will readily plug-in to Slicer3.
- BRAINSFit - Linear fitting of two 3D image sets using a mutual information similarity metric
- BRAINSDemonsWarp - Non-linear registration of two image sets. This includes the diffeomorphic demons algorithm along with a number of useful enhancements.
- BRAINSMush - Tool to combine T1 and T2 datasets into a composite image. The resulting image is then used to perform brain extraction.
- Neural Networks - Automated segmentation of subcortical and cerebellar regions of interest using an artificial neural network.
- BRAINSTracer - Tracing tool that uses thge vtkContourWidget for defining a region of interest manually
- GTRACT - Diffusion tensor analysis and Fiber tracking tool
- BRAINS Auto Workup - Fully automated image analysis pipeline for generation of volumetric measurements.
BRAINS Features
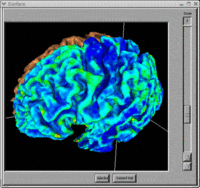
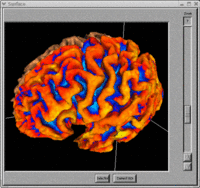
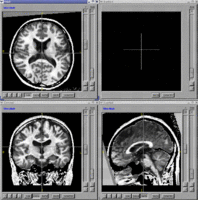
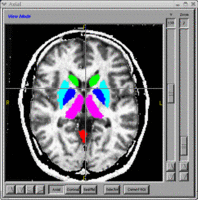
- BRAINS Gallery
Publications
- Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. Neuroimage. 2007 Aug 22; [Epub ahead of print].