Difference between revisions of "Projects:DBP2:Harvard:Registration Documentation"
| Line 5: | Line 5: | ||
Talk to Andrew Rausch, rauscha@bwh.harvard.edu for more info. I'm no expert on registration, but I can tell you exactly how each of these were performed and provide mrmls and nrrds. | Talk to Andrew Rausch, rauscha@bwh.harvard.edu for more info. I'm no expert on registration, but I can tell you exactly how each of these were performed and provide mrmls and nrrds. | ||
| + | |||
| + | =Special case: Anna's data registration= | ||
=There and back registration= | =There and back registration= | ||
Revision as of 14:03, 4 May 2009
Home < Projects:DBP2:Harvard:Registration DocumentationSlicer 3 contains a number of different ways to register two images. In this document we will examine the results of these modules on three different cases. The convention used will be (moving image) -> (fixed image) = (resulting moved image). The cases are:
- Registering a t2 weighted image to an arbitrary t1, then registering this transformed image back to the original t2. Ideally this will produce a identical image.
- Registering a DTI baseline image to the corresponding realigned t2 weighted image, and vice-versa. This is a realistic test of one of our (the PNL) everyday uses of registration.
- Registering a realigned t2 weighted image to another t2.
Talk to Andrew Rausch, rauscha@bwh.harvard.edu for more info. I'm no expert on registration, but I can tell you exactly how each of these were performed and provide mrmls and nrrds.
Contents
Special case: Anna's data registration
There and back registration
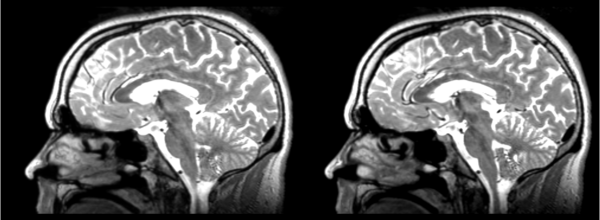
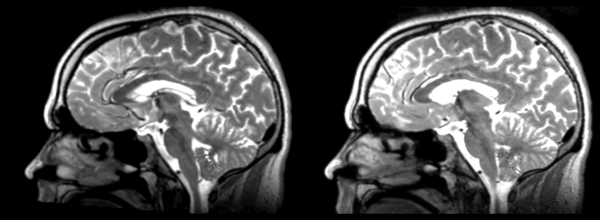
In this case we take the 01053-t2w nrrd as our moving image and the 01031-t1 nrrd as our fixed, register them and save the result, then move that result to the fixed 01053-t2w image. Not all of these transformations are totally reversible, but for the most part if they can be registered one way, they should be able to be registered back to exactly the same spot. It never works quite perfectly though, so the errors/distortions are detailed below.
Slicer modules
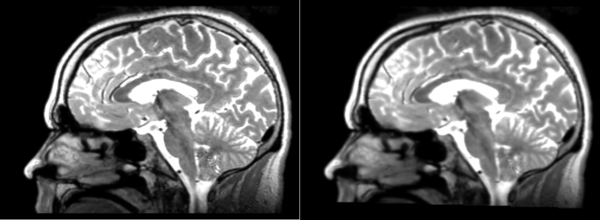
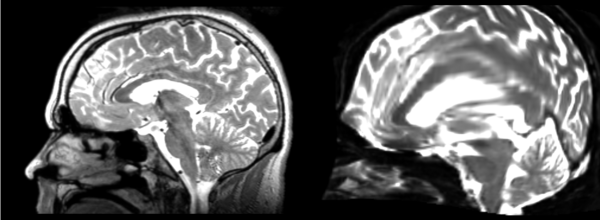
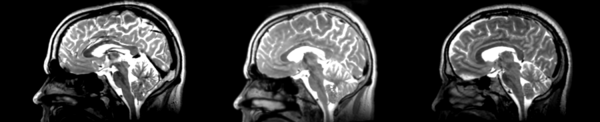
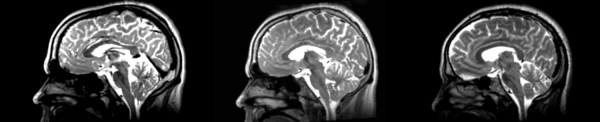
Affine
- 53-t2 -> 31-t1 = aff1
- aff1 -> 53-t2 = aff2
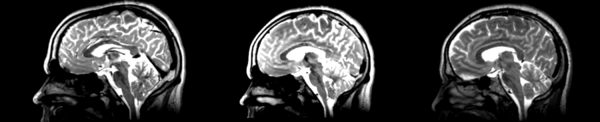
The image shows these side by side. The are nearly the same except for the loss of fine detail.
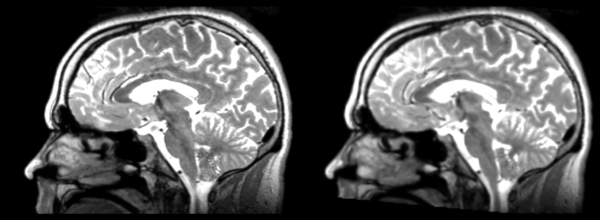
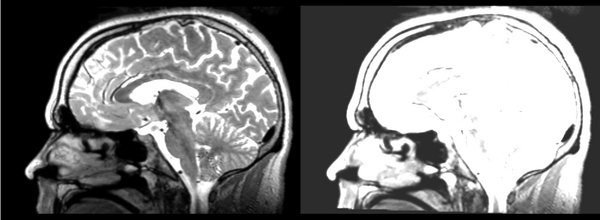
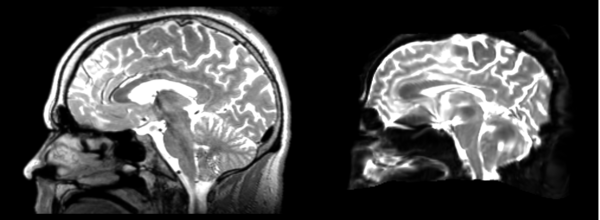
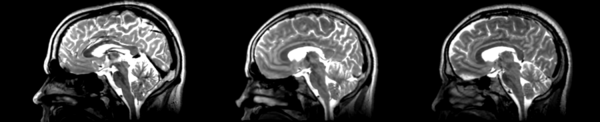
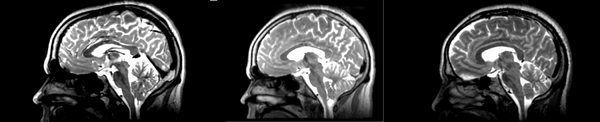
Linear
- 53-t2 -> 31-t1 = lin1
- lin1 -> 53-t2 = lin2
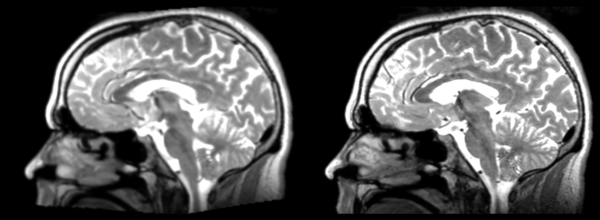
The image shows lin2 and 53-t2 side by side. The image shows there appears to be some change beyond loss of detail, possibly a L-R shift in the front.
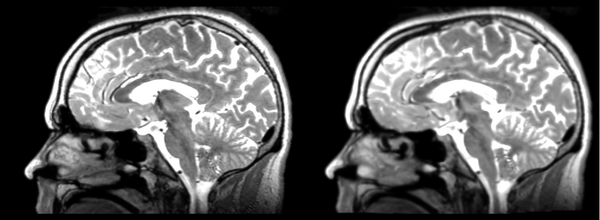
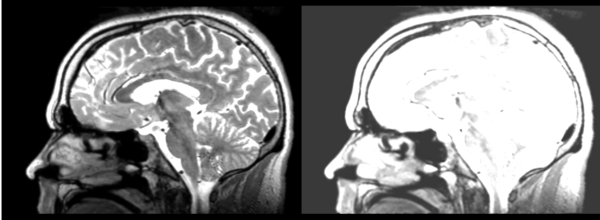
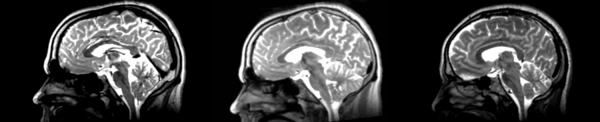
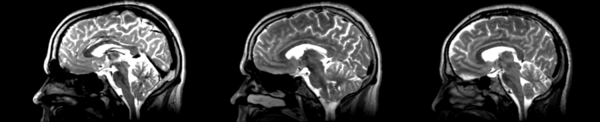
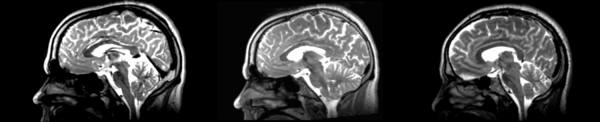
Rigid
- 53-t2 -> 31-t1 = rig1
- rig1 -> 53-t2 = rig2
Significant blurring, but every feature is in it's proper place.
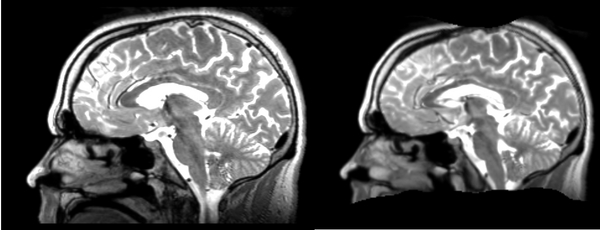
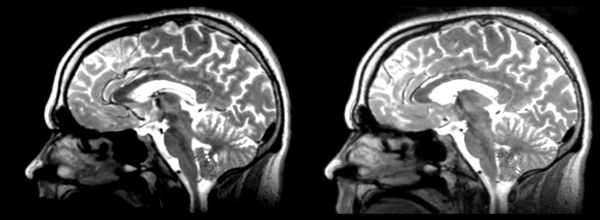
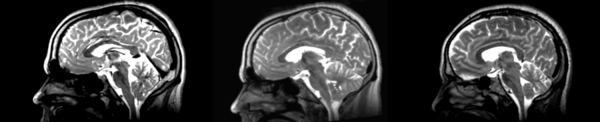
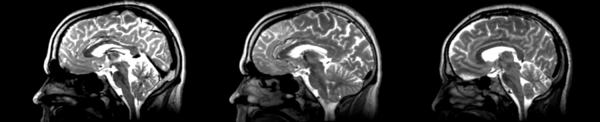
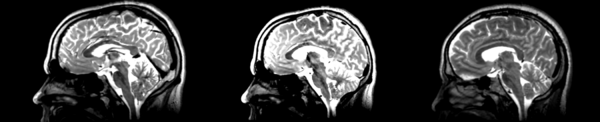
Deformable B-spline
- 53-t2 -> 31-t1 = Bspline1
- Bspline1 -> 53-t2 = Bspline2
Superior frontal enlargement, odd upward shifting in rear of brain, distortions everywhere.
Diffeomorphic Demons
- 53-t2 -> 31-t1 = Demons1
- Demons1 -> 52-t2 = Demons2
Multiple deformations. Much larger ventricles. Small errors everywhere, expansions, contractions, shifts. Roughly in the same place overall.
Register Images Module
Initial
- 53-t2 -> 31-t1 = init1
- init1 -> 53-t2 = init2
Significant shift in count values, rigid translation down ~4 pixels, left ~1 pixel.
Rigid
- 53-t2 -> 31-t1 = rigid1
- rigid1 -> 53-t2 = rigid2
Shift in grey values. Few other problems.
Affine
- 53-t2 -> 31-t1 = affine1
- affine1 -> 53-t2 = affine2
Again, shift in grey values, mostly okay otherwise. Slight posterior shift.
B-Spline
- 53-t2 -> 31-t1 = Bspline1
- Bspline1 -> 53-t2 = Bspline2
Again grey values shift. Minor distortion in frontal lobe and cerebellum.
Pipe Rigid
- 53-t2 -> 31-t1 = piperigid1
- piperigid1 -> 53-t2 = piperigid2
The brain appears to be shifted slightly leftward, as the contours around the edges do not perfectly match. There also is a tilt forward around the L-R axis. Grey value shift.
Pipe Affine
- 53-t2 -> 31-t1 = pipeaffine1
- pipeaffine1 -> 53-t2 = pipeaffine2
Elongation along the superior frontal to cerebellum axis. Grey value shift. Otherwise ok.
Pipe Bspline
- 53-t2 -> 31-t2 = pipebspline1
- pipebspline1 -> 53-t2 = pipebspline2
There is pinching towards the corpus callosum, strange distortions near the cerebellum, many distortions around the skull, very poor overall.
.
DTI Baseline to t2 registration
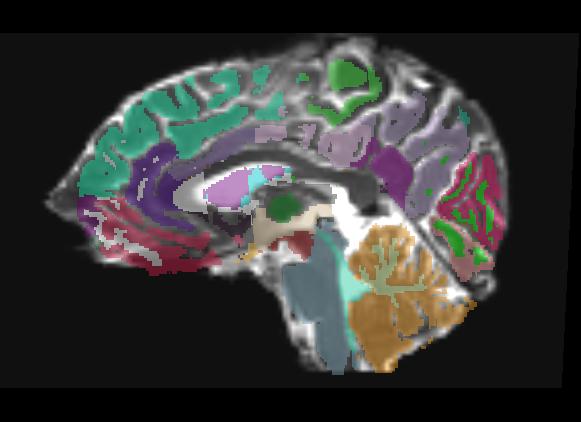
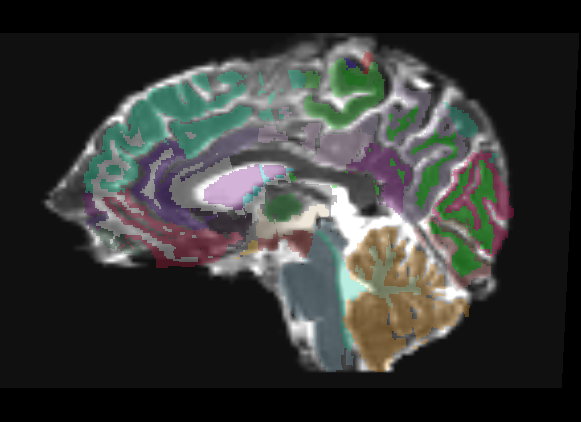
Freesurfer parcellation overlay
Only linear methods produce transforms that can be used to register a label map. Unfortunately, a linear registration can never account for the distortions of a DTI image, and as such there is no way to get a good registration of structural label maps to DTI space. In Slicer 2, I can create a transform node and do an automatic registration between two already created nrrds based on intensity via diff. demons or bspline, and then apply that transform to a label map, but I don't know of any way to get a similar transform in Slicer3.
Affine
The overlay has very few places where it is even usable, only in the dead center of ROIs do you have any guarantee of being in the same region on the DTI. This is only a linear registration, so the label map cannot account for the DTI distortion.
Linear
The overlay has very few places where it is even usable, only in the dead center of ROIs do you have any guarantee of being in the same region on the DTI. This is only a linear registration, so the label map cannot account for the DTI distortion.
Slicer modules
B-Spline
- DTI-baseline -> 53-t2 = Bspline3
Fits the cortex on the baseline to the skull on the t2. Retry with a skull stripped t2.
Diffeomorphic Demons
- DTI-baseline -> 53-t2 = Demons3
Different subject registration
Slicer modules
Rigid
01053>01031rigid.png = not bad... different brains of different sizes, but seem to be aligned in the ventricles so output seems okay. output slightly blurry. bad initial leveling displays.
Linear
01053>01031linear.png = not bad... different brains of different sizes, but seem to be aligned in the ventricles so output seems okay.output slightly blurry. bad initial leveling displays.
Affine
01053>01031_affine.png = looks good in terms of size. odd warping of frontal lobe and cerebellum area, but otherwise good coregistration.
B-spline
01053>01031_bspline.png = looks good in terms of size. odd warping of frontal/temporal lobe and cerebellum area, but otherwise good coregistration.
Diffeomorphic Demons
01053>01031_demons.png = 10 minutes. Window/leveling initially off. Otherwise, it looks really good. Not much distortion.
Register Images module
Initial
01053>01031_RI_initial.png - looks like the original 01053 image, but shifted, which is expected. resolultion a little worse.
Rigid
01053>01031_RI_rigid.png - center ventricles are mostly aligned, but not perfectly. in axial view, brain is more crooked than in initial scan... especially in frontal lobe.
Affine
01053>01031_RI_affine.png - ventricles aligned well. sizing decent. looks how one would expect affine to look. not perfect subcortically. some distortion, esp in temporal lobe.
B-spline
01053>01031_RI_bspline.png - good result. ventricles aligned well, but temporal lobe size is not the same as the target image.
Pipe Rigid
01053>01031_RI_pipe_rigid.png - shifted version of the original image. decent registration in the ventricles.
Pipe Affine
01053>01031_RI_pipe_affine.png - decent coreg. a bit of distortion in the frontal and temporal lobes, esp around the corpus collosum.
pipe B-spline
01053>01031_RI_pipe_bspline.png - good coregistration, but pretty blurred. also, I would have expected the temporal lobe to be coregistered better.