Difference between revisions of "2009 Summer Project Week Slicer3 XNAT usecases"
| Line 1: | Line 1: | ||
[[image:XNAT_IGTUseCase.png]] | [[image:XNAT_IGTUseCase.png]] | ||
| + | |||
| + | |||
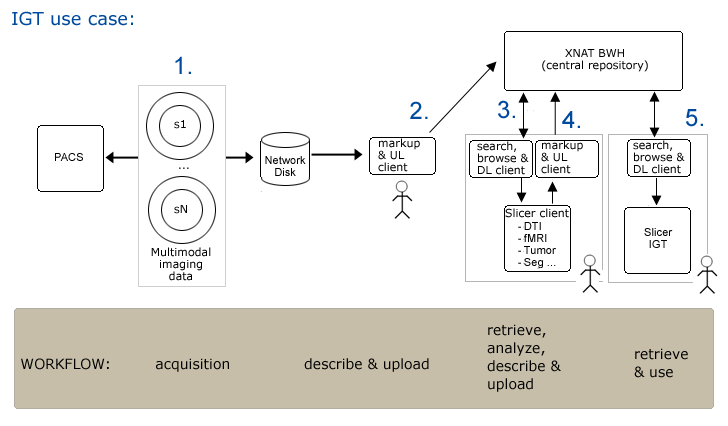
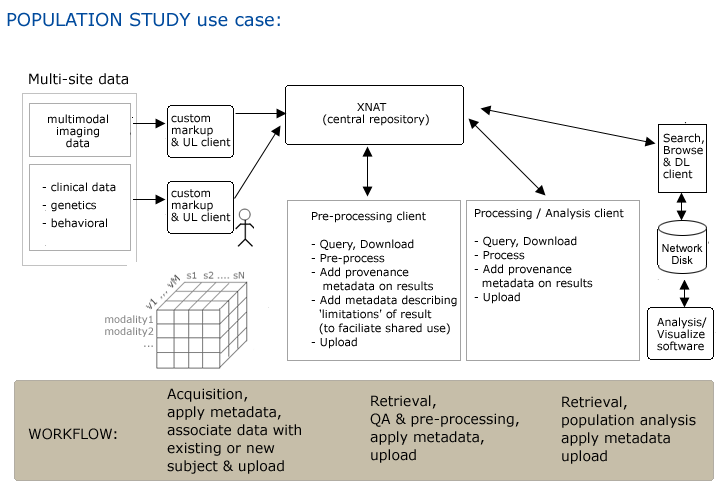
| + | '''Step1.''' Subject is scanned and data is pushed to a networked disk (and referenced by a PACS system) automatically. | ||
| + | |||
| + | '''Step2.''' Researcher1 uses an XND client (on a laptop or a desktop system) to mark up subject data and to upload it (via drag and drop) to the central XND repository xnd.bwh.harvard.edu. | ||
| + | |||
| + | '''Step3.''' Researcher2 opens slicer and queries XND webservices on svr = http://xnd.bwh.harvard.edu:8081 for a list of subjects. | ||
| + | |||
| + | Query for all subjects on system... | ||
| + | '''send:''' curl $srv/search??subject | ||
| + | '''response:''' xml for all subjects in xnd.bwh.harvard.edu | ||
| + | |||
| + | Choose a subject, query for available data... | ||
| + | '''send:''' curl $srv/search?subject=$subj | ||
| + | '''response:''' xml containing URIs of all resources for the individual subject $subj | ||
| + | |||
| + | Slicer parses XML and presents list of all resources; researcher2 queries for for only DICOM data for a particular subject... | ||
| + | '''send:''' curl $srv/search?subject=$subj&file_type=DICOM | ||
| + | '''response:''' xml containing URIs of all DICOM resources for subject $subj | ||
| + | |||
| + | Slicer parses XML and presents list of URIs, researcher2 requests to download uris of interest... | ||
| + | '''send:''' curl $uri | ||
| + | '''response: file contents | ||
| + | |||
| + | '''Step4.''' Completed neurosurgical planning analysis and upload results. | ||
| + | |||
| + | Create metadatadocument (XML) for individual data file, or for scene + selected data. First, post each dataset, and assemble MRML file using returned uris. Then, post or put mrml file. | ||
| + | '''send:''' curl -X POST -H "Content-Type: application/x-xnat-metadata+xml" -H "Content-Disposition: x-xnat-metadata; filename=\"$filename\"" -d @$metadata.xml $srv/data | ||
| + | '''response:''' URI of posted file (as message body, also in Location: HTTP header) | ||
| + | '''send:''' curl -T $filepath $url | ||
| + | '''response:''' none | ||
| + | |||
| + | '''Step5.''' In IGT, download scene for patient (subject) into slicer. | ||
| + | |||
| + | Query for a subject and query for available scene files... | ||
| + | '''send:''' curl $srv/search?subject=$subj&fileType=MRML | ||
| + | '''response:''' xml containing URIs of all MRML files | ||
| + | |||
| + | Slicer parses XML and presents list of MRML files, and clinican selects one with $uri. | ||
| + | '''send:''' curl -T $local_file $uri | ||
| + | '''response:''' none | ||
| + | |||
| + | Clinician cuts network link to remote repository (all files are locally cached so OR system is self sufficient). | ||
[[image:XNAT_RTPlanningUseCase.png]] | [[image:XNAT_RTPlanningUseCase.png]] | ||
Revision as of 10:40, 24 June 2009
Home < 2009 Summer Project Week Slicer3 XNAT usecases
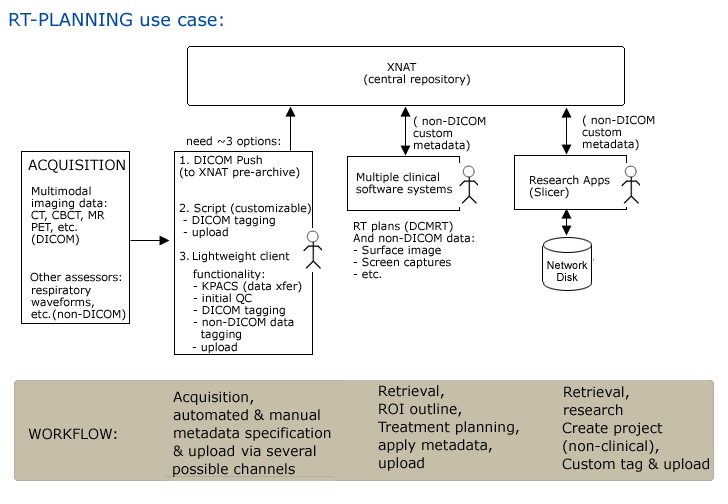
Step1. Subject is scanned and data is pushed to a networked disk (and referenced by a PACS system) automatically.
Step2. Researcher1 uses an XND client (on a laptop or a desktop system) to mark up subject data and to upload it (via drag and drop) to the central XND repository xnd.bwh.harvard.edu.
Step3. Researcher2 opens slicer and queries XND webservices on svr = http://xnd.bwh.harvard.edu:8081 for a list of subjects.
Query for all subjects on system...
send: curl $srv/search??subject response: xml for all subjects in xnd.bwh.harvard.edu
Choose a subject, query for available data...
send: curl $srv/search?subject=$subj response: xml containing URIs of all resources for the individual subject $subj
Slicer parses XML and presents list of all resources; researcher2 queries for for only DICOM data for a particular subject...
send: curl $srv/search?subject=$subj&file_type=DICOM response: xml containing URIs of all DICOM resources for subject $subj
Slicer parses XML and presents list of URIs, researcher2 requests to download uris of interest...
send: curl $uri response: file contents
Step4. Completed neurosurgical planning analysis and upload results.
Create metadatadocument (XML) for individual data file, or for scene + selected data. First, post each dataset, and assemble MRML file using returned uris. Then, post or put mrml file.
send: curl -X POST -H "Content-Type: application/x-xnat-metadata+xml" -H "Content-Disposition: x-xnat-metadata; filename=\"$filename\"" -d @$metadata.xml $srv/data response: URI of posted file (as message body, also in Location: HTTP header) send: curl -T $filepath $url response: none
Step5. In IGT, download scene for patient (subject) into slicer.
Query for a subject and query for available scene files...
send: curl $srv/search?subject=$subj&fileType=MRML response: xml containing URIs of all MRML files
Slicer parses XML and presents list of MRML files, and clinican selects one with $uri.
send: curl -T $local_file $uri response: none
Clinician cuts network link to remote repository (all files are locally cached so OR system is self sufficient).