Difference between revisions of "Projects:RegistrationLibrary:RegLib C02"
From NAMIC Wiki
m (→Procedures) |
|||
| Line 46: | Line 46: | ||
*download/view guided video tutorial | *download/view guided video tutorial | ||
*download power point tutorial | *download power point tutorial | ||
| + | {| style="cellpadding="10" cellspacing="0" border="0" | ||
| + | |Load Transform | ||
| + | |none | ||
| + | |- | ||
| + | |Save Transform: | ||
| + | |Create New Linear Transform | ||
| + | |- | ||
| + | | Initialization: | ||
| + | |none | ||
| + | |- | ||
| + | | Registration: | ||
| + | | Affine | ||
| + | |} | ||
| − | + | <br> | |
| − | |||
| − | |||
Registration: Affine | Registration: Affine | ||
Metric: NormCorr | Metric: NormCorr | ||
Revision as of 19:52, 20 October 2009
Home < Projects:RegistrationLibrary:RegLib C02Contents
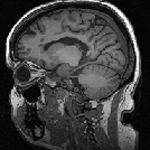
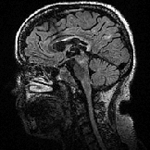
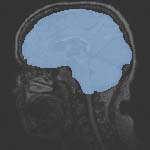
Slicer Registration Use Case Exampe: Intra-subject Brain MR FLAIR to MR T1
|
|
| |
| 1mm isotropic 256 x 256 x 146 RAS |
1.2mm isotropic 256 x 256 x 116 RAS |
1.2mm isotropic 256 x 256 x 116 RAS |
Objective / Background
This scenario occurs in many forms whenever we wish to align all the series from a single MRI exam/session into a common space. Alignment is necessary because the subject likely has moved in between series.
Keywords
MRI, brain, head, intra-subject, FLAIR, T1, defacing, masking, labelmap, segmentation
Input Data
- reference: T1 SPGR , 1x1x1 mm voxel size, sagittal, RAS orientation
- moving: T2 FLAIR 1.2x1.2x1.2 mm voxel size, sagittal, RAS orientation
- Content preview: Have a quick look before downloading: Does your data look like this? SPGR Lighbox , FLAIR Lighbox
- download dataset to load into slicer (11.4 MB zip archive)
Registration Challenges
- we expect the amount of misalignment to be small
- we know the underlying structure/anatomy did not change, hence whatever residual misalignment remains is of technical origin.
- the different series may have different FOV. The additional image data may distract the algorithm and require masking
- the different series may have very different resolution and anisotropic voxel sizes
- hi-resolution datasets may have defacing applied to one or both sets, and the defacing-masks may not be available
- the different series may have different contrast.
- individual series may contain motion or other artifacts
Key Strategies
- the SPGR is the anatomical reference. It is also higher resolution. Unless there are overriding reasons, always use the highest resolution image as your fixed/reference.
- the defacing of the SPGR image introduces sharp edges that can be detrimental. Best to mask that area. If you have the mask available, use it. But in this case since we already have a skull-stripping mask as part of the labelmap, that is even better. We will load the labelmap and use it as mask in finding the registration
- because the two images are still reasonably similar in contrast, we can choose an intensity ratio as cost function, which is less stable but if successful provides a more precise alignment than mutual information.
Procedures
- download step-by step text instructions
- download recommended parameter settings here
- download/view guided video tutorial
- download power point tutorial
| Load Transform | none |
| Save Transform: | Create New Linear Transform |
| Initialization: | none |
| Registration: | Affine |
Registration: Affine
Metric: NormCorr
Expected offset magnitude: 10
Expected rotation magnitude: 0.1
Expected scale magnitude: 0.05
Expected skew magnitude: 0.01
Verbosity Level: standard
Fixed Image Mask: S021_ICC
Random Number Seed: 0 (none)
Number of threads: 0 (max)
Interpolation: linear
Affine Max Iterations: 50
Affine Sampling Ratio: 0.02
Registration Results
- registration parameter presets file (load into slicer and run the registration)
- result transform file (load into slicer and apply to the target volume)
- result screenshots (compare with your results)
- result evaluations (metrics)