Difference between revisions of "NA-MIC/Projects/Structural/Segmentation/Rule based segmentation:Striatum"
m (Update from Wiki) |
|||
| Line 17: | Line 17: | ||
* [[DBP:Harvard:Collaboration:GTech|Harvard - Rule-based Segmentation]] | * [[DBP:Harvard:Collaboration:GTech|Harvard - Rule-based Segmentation]] | ||
| − | * [[Algorithm: | + | * [[Algorithm:Stony Brook#Rule_Based_Segmentation_Slicer_Modules||Stony Brook Summary Page]] |
Latest revision as of 01:10, 16 November 2013
Home < NA-MIC < Projects < Structural < Segmentation < Rule based segmentation:StriatumBack to NA-MIC_Collaborations
Objective: Semi Automatic Segmentation and parcellation for the Basal Ganglia.
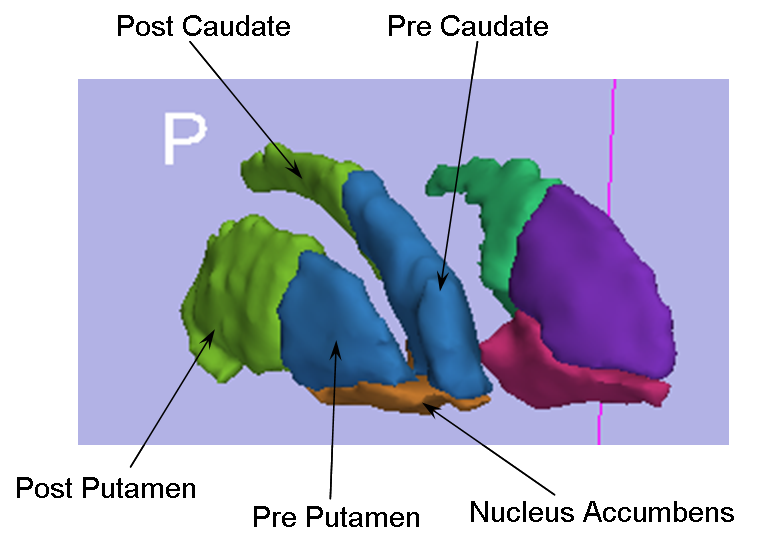
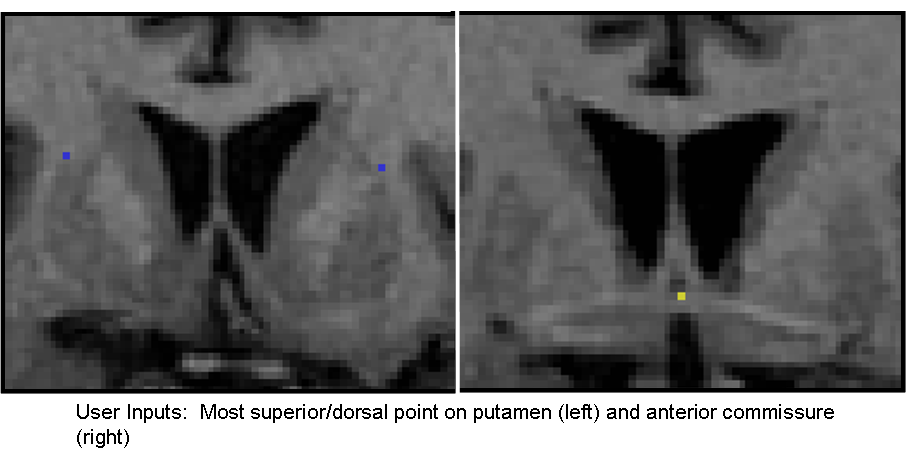
Progress: We have developed an algorithm for delineation of the striatum into 5 physiological subregions (pre/post caudate, pre/post putamen, and nucleus accumbens) while requiring only minimal user input. We have implemented this algorithm from the geometric rules for delineating the striatum as defined by our Core 3 collaborator, Dr. James Levitt of the PNL, into a 3D SLICER module. The current run time for the algorithm is ~20 seconds after the initial user input. The user inputs a label map of the full striatum, the most superior/dorsal voxel of the putamen on each slice, and the anterior commisure voxel (see figure below). From these, the labelmap is delineated into the aforementioned subregions. The figure below shows a 3D model of the left and right striatum delineated into the five subregions.
Key Investigators:
- GTech: Ramsey Al-Hakim, Delphine Nain, Allen Tannenbaum.
- PNL: Sylvain Bouix, James Levitt, Marc Niethammer, Martha Shenton.
- Kitware: Luis Ibanez
- Isomics: Steve Pieper
Links: