Difference between revisions of "RobustStatisticsSegmentation"
| Line 157: | Line 157: | ||
The method is based on: | The method is based on: | ||
| − | * [http://www.na-mic.org/publications/pages/display?search=StatisticalSegmentationSlicer2&submit=Search&words=all&title=checked&keywords=checked&authors=checked&abstract=checked&searchbytag=checked&sponsors=checked| NA-MIC Publications Database] | + | * [http://www.na-mic.org/publications/pages/display?search=StatisticalSegmentationSlicer2&submit=Search&words=all&title=checked&keywords=checked&authors=checked&abstract=checked&searchbytag=checked&sponsors=checked| NA-MIC Publications Database on Simultaneous Multiple Object Segmentation using Robust Statistics Features] |
* Yang, Y. and Tannenbaum, A. and Giddens, D. and Coulter, WH, "Knowledge-based 3D segmentation and reconstruction of coronary arteries using CT images", in IEEE EMBS 2004, pp1664--1666 | * Yang, Y. and Tannenbaum, A. and Giddens, D. and Coulter, WH, "Knowledge-based 3D segmentation and reconstruction of coronary arteries using CT images", in IEEE EMBS 2004, pp1664--1666 | ||
==Rons== | ==Rons== | ||
[[File:RobustSeg.zip|example case]] | [[File:RobustSeg.zip|example case]] | ||
Revision as of 19:42, 11 May 2010
Home < RobustStatisticsSegmentationContents
- 1 Robust Statistics Based Segmentation
- 1.1 Description
- 1.2 How to get the module
- 1.3 Usage
- 1.4 Testing
- 1.5 Testing case: left kidney
- 1.6 Testing case: right kidney
- 1.7 Testing case: spleen
- 1.8 Testing case: liver
- 1.9 Testing case: kidneys, spleen, liver
- 1.10 Testing case: tumor
- 1.11 Testing case: tumor
- 1.12 Testing case: tumor
- 1.13 Testing case: vervet brain
- 1.14 Key Investigators
- 1.15 Publications
- 1.16 Rons
Robust Statistics Based Segmentation
Description
Given a few initial seeds, we use the robust statistics to construct a feature image out of the original image, and the image segmentation is carried out in the feature image. To do so, we first compute the certain robust statistics, such as the median absolute deviation(MAD) and the interquartile range (IQ), from the seed points. Further, a multi-dimentional probability distribution function is computed from the feature seeds. At each point in the image, the active contour is driven by the probabilities of this pixel belonging to each category indicated by the seeds.
How to get the module
- start nightly built Slicer 3.5 (Most recently tested is Slicer3-3.5-alpha-2010-01-11-linux-x86_64)
- open View->Extension Manager
- check "Find & Install", click "Next"
- in the list, select "RobustStatisticsSegmentor", then "Download & Install", and "Next"
- Slicer will ask to restart, confirm.
- after restarts, the module is in Module category: "Segmentation-->Robust Statistics Segmentation"
Usage
General guideline
- First run:
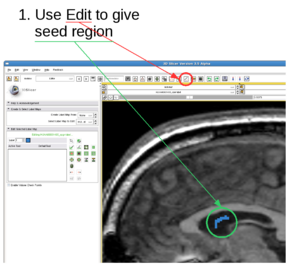
- Give a rough estimate of the object volume and put fiducial points (preferred to be 2 or more) in the object.
- Run the module using the default parameters.
- Note:
- The Approximate volume is just a rough upper limit for the volume. It should be at least the size of the object. This is because when the volume reaches that, the program must stop. However, other criteria may stop the algorithm before the volume reaches this value.
- The fiducial points can be thrown into the object. What I do is I just add two fiducial points and move them into the object within one slice.
- Troubleshooting
- Surface is too rough. Try:
- Increase "Boundary smoothness"
- Leakage into thin/narrow regions. Try:
- Increase "Boundary smoothness"
- leakage into similar (but still different) intensity regions (which is not necessarily thin), Try:
- Increase "Intensity homogeneity"
- Some regions are missed: Try (either one):
- Increase "Max volume"
- Decrease"Intensity homogeneity"
- Decrease "Boundary smoothness"
- Some regions are missed, at the same time leakages to some other regions. Try (either one)
- Increase "Intensity homogeneity"
- Add another fiducial point
- Surface is too rough. Try:
Multiple-value label map handeling
The parameter "Output Label Value"(OLV) is for user to assign the output label value. Moreover, it is also used in cases where the user provided label map contains more than one label value. In general, the module handles three cases:
- user provided label map contains only one label value, L. In this case, the output label value will be set to OLV, not matter what value L takes.
- user provided label map contains multiple label values, one of which matches OLV. Then only that label will be effective and all the others are discarded. The output will have label value OLV too.
- user provided label map contains multiple label values, but none matches OLV. Then all the non-zero labels will be considered as a single label value and then come back to the case 1 above.
Testing
Several tests are conducted and shown here along with the parameters used to get the results.
Testing case: left kidney
- Data set: http://wiki.na-mic.org/Wiki/images/8/8d/Patient1.tar.gz
- Approximate volume: 200 mL
- Intensity homogeneity: 0.1
- Boundary smoothness: 0.5
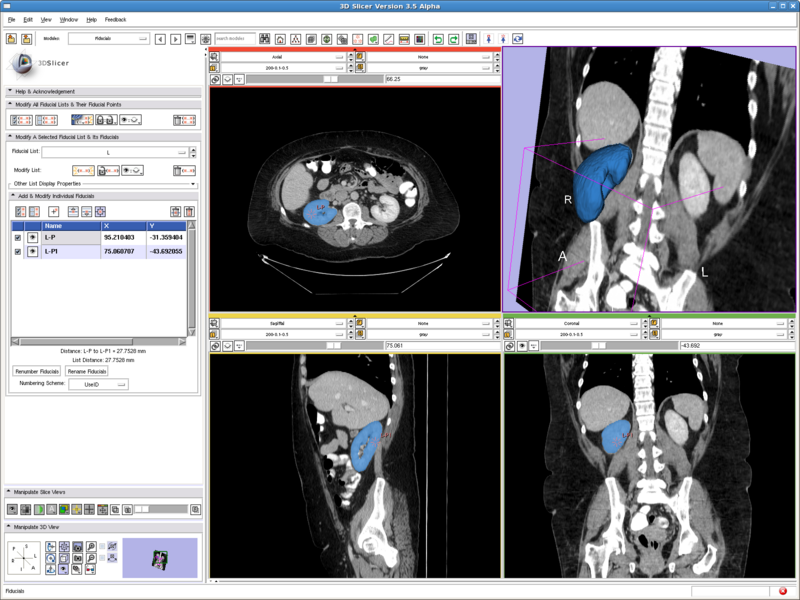
Testing case: right kidney
- Data set: http://wiki.na-mic.org/Wiki/images/8/8d/Patient1.tar.gz
- Approximate volume: 200 mL
- Intensity homogeneity: 0.1
- Boundary smoothness: 0.5
In the figure below we show the position of the two seeds. They are both in the cortex region. And it can be seen in the segmentation result, the renal cortex got segmented and the pelvis is not.
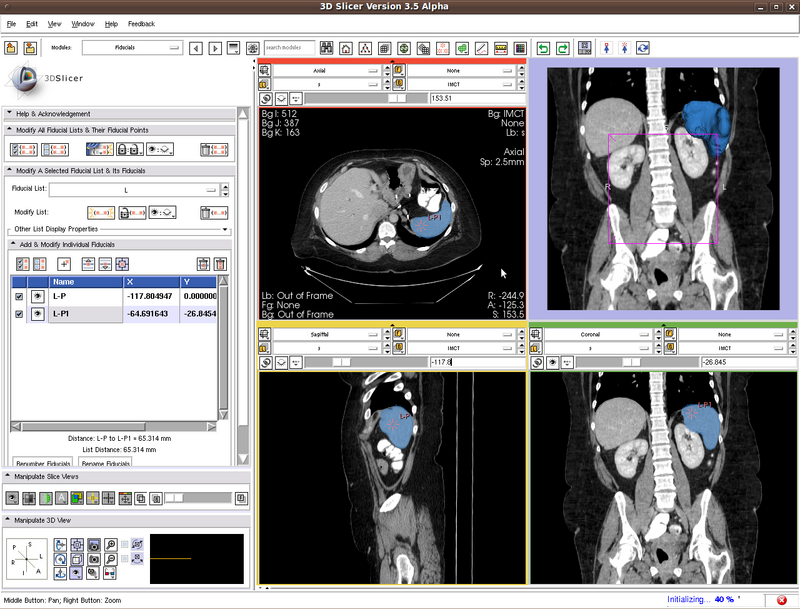
Testing case: spleen
- Data set: http://wiki.na-mic.org/Wiki/images/8/8d/Patient1.tar.gz
- Approximate volume: 300 mL
- Intensity homogeneity: 1
- Boundary smoothness: 0.3
In the figure below we show the position of the two seeds.
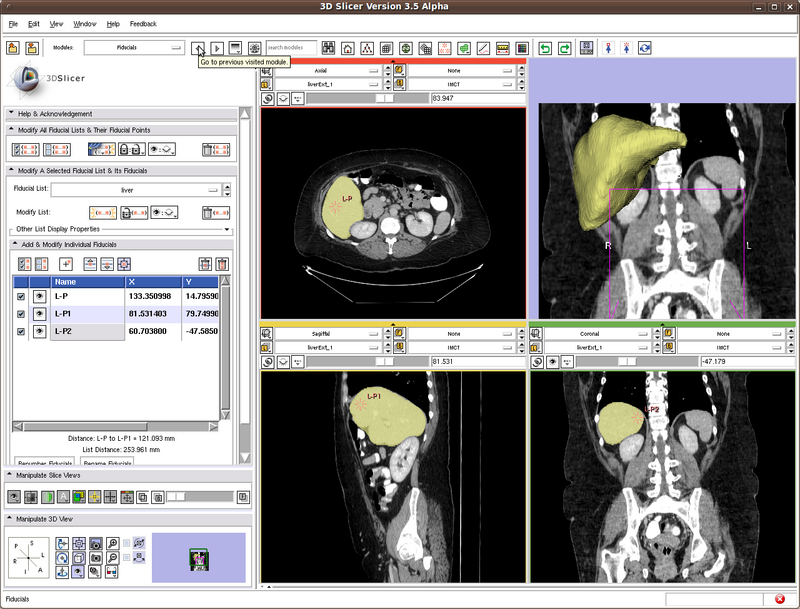
Testing case: liver
- Data set: http://wiki.na-mic.org/Wiki/images/8/8d/Patient1.tar.gz
- Approximate volume: 2000 mL
- Intensity homogeneity: 0.8
- Boundary smoothness: 0.8
- Max running time (min): 20
In the figure below we show the position of the three seeds.
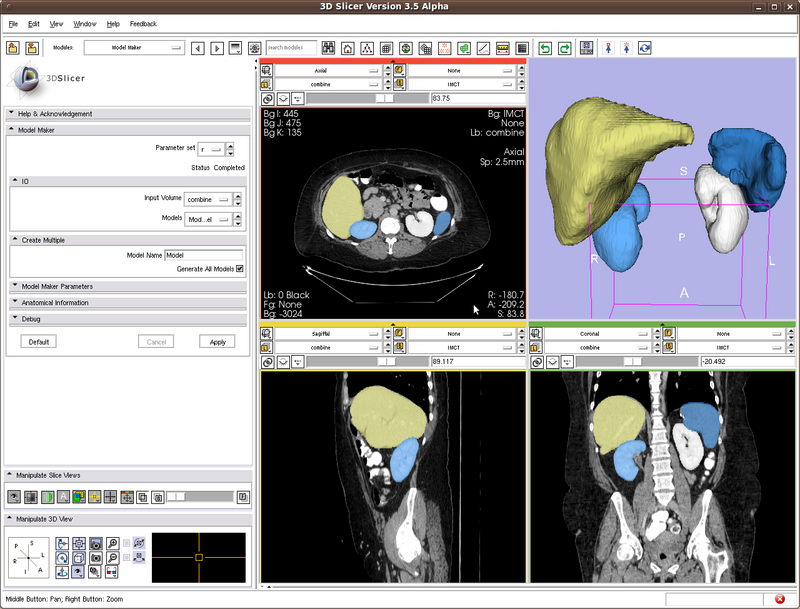
Testing case: kidneys, spleen, liver
User can chose output label value. If different labels are used for different organs, then user can use the "Modules->Filtering->Image Label Combine" to combine the different label maps into one. This is shown in the following figure.
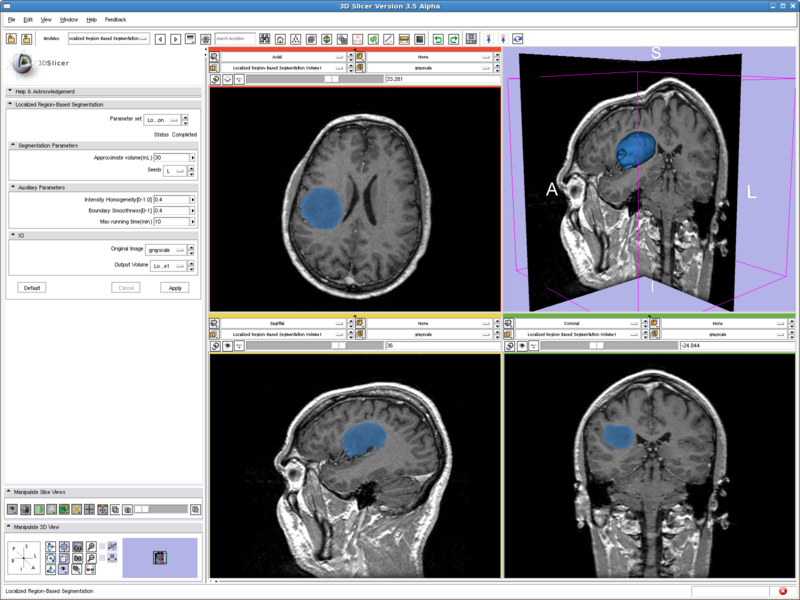
Testing case: tumor
- Data set: http://wiki.na-mic.org/Wiki/images/0/0f/MayExperiments.zip (DiffusionEditorBaselineNode.nrrd)
- Approximate volume: 30 mL
- Intensity homogeneity: 0.1
Testing case: tumor
- Data set: http://www.spl.harvard.edu/publications/bitstream/download/4217 (case3/grayscale.nrrd)
- Approximate volume: 30 mL
- Intensity homogeneity: 0.4
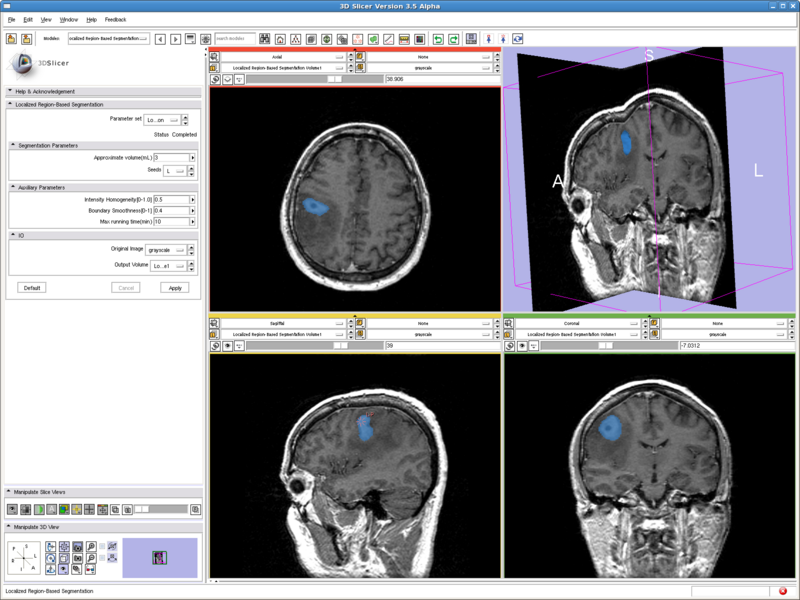
Testing case: tumor
- Data set: http://www.spl.harvard.edu/publications/bitstream/download/4217 (case5/grayscale.nrrd)
- Approximate volume: 3 mL
- Intensity homogeneity: 0.5
- Boundary smoothness: 0.4
This is a difficult case. All the other methods tested either captures only the middle dark spot, or leaks out of the tumor. But the method here nicely capture both the dark core and the bright shell, without leaking out to regions with intensities in the middle.
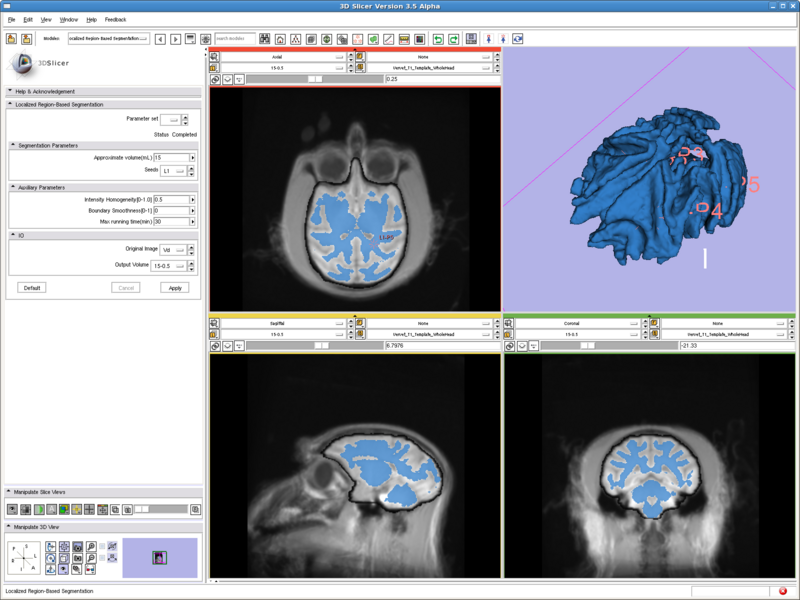
Testing case: vervet brain
- Data set: http://www.na-mic.org/Wiki/index.php/Vervet_MRI_registration (http://www.bsl.ece.vt.edu/data/vervet_atlas/VPA-10-1.1.zip Vervet_T1_Template_WholeHead.nii)
- Approximate volume: 15 mL
- Intensity homogeneity: 0.5
Previous tests all run with two fiducial points, and running time are all less than 10 seconds. This case has 6 points and runs for 15min to get the result shown.
Key Investigators
Georgia Tech: Yi Gao and Allen Tannenbaum
BWH: Katie Hayes, Andriy Fedorov, and Ron Kikinis
Publications
The method is based on:
- NA-MIC Publications Database on Simultaneous Multiple Object Segmentation using Robust Statistics Features
- Yang, Y. and Tannenbaum, A. and Giddens, D. and Coulter, WH, "Knowledge-based 3D segmentation and reconstruction of coronary arteries using CT images", in IEEE EMBS 2004, pp1664--1666