Difference between revisions of "Projects:RegistrationTestbed"
Jfishbaugh (talk | contribs) |
Jfishbaugh (talk | contribs) |
||
| Line 22: | Line 22: | ||
==Registration Testbed== | ==Registration Testbed== | ||
| + | |||
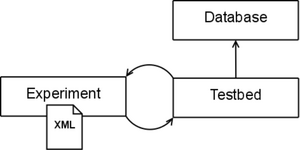
| + | Our proposed framework is similar in spirit to that of [http://www.batchmake.org Batchmake]. At the highest level of abstraction, the testbed is a common interface to command line registration applications. It allows for batch processing large experiments for single or multiple registration pipelines. We use the term pipeline to describe a collection of registration applications that are associated with each other. For example, the [http://www.doc.ic.ac.uk/~dr/software/ IRTK] pipeline consists of three applications: one for linear registration, one for non-linear registration, and finally an application to produce the output image. While there are three separate applications, they are used together for a single registration task. A pipeline in our testbed consists of one or more registration applications. | ||
[[Image:Diagram1.png|thumb|center|300px|High level architecture.]] | [[Image:Diagram1.png|thumb|center|300px|High level architecture.]] | ||
| − | |||
| − | |||
At a high level, the testbed has three main components. First, the database represents a collection of definitions of known registration pipelines. This includes knowledge of required and optional input parameters and format of external parameter files used as input. The definitions allow for the implicit enforcement of any rules a given application might have. For example, an application might require a target and source image as the first two arguments, or optional parameters be specified with a "-" or "--". In other words, the database of known definitions helps distance a user from application specific details. This way, a user can more easily generate experiments with one or more registration pipelines without having to know how to use that specific application or consult various documentation. Any command line based registration application can be easily added to the database. | At a high level, the testbed has three main components. First, the database represents a collection of definitions of known registration pipelines. This includes knowledge of required and optional input parameters and format of external parameter files used as input. The definitions allow for the implicit enforcement of any rules a given application might have. For example, an application might require a target and source image as the first two arguments, or optional parameters be specified with a "-" or "--". In other words, the database of known definitions helps distance a user from application specific details. This way, a user can more easily generate experiments with one or more registration pipelines without having to know how to use that specific application or consult various documentation. Any command line based registration application can be easily added to the database. | ||
| Line 31: | Line 31: | ||
Experiments are stored in an XML format. An experiment file consists of necessary experiment information, such as paths to executables and parameter values. The main testbed application receives an experiment file as input, and using the database, formats the correct command line calls to conduct the experiment. An advanced user can interact directly with the XML to develop experiments. However, a user interface is available to help automate the process. | Experiments are stored in an XML format. An experiment file consists of necessary experiment information, such as paths to executables and parameter values. The main testbed application receives an experiment file as input, and using the database, formats the correct command line calls to conduct the experiment. An advanced user can interact directly with the XML to develop experiments. However, a user interface is available to help automate the process. | ||
| − | [[Image:Xmlexample.png|thumb|300px|A simple experiment file.]] | + | [[Image:Xmlexample.png|thumb|center|300px|A simple experiment file.]] |
===Examples=== | ===Examples=== | ||
| Line 39: | Line 39: | ||
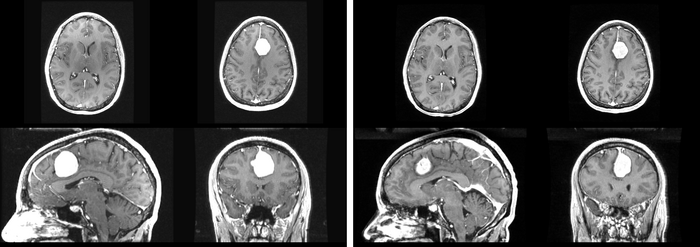
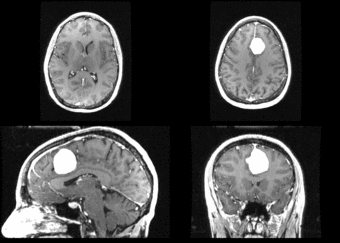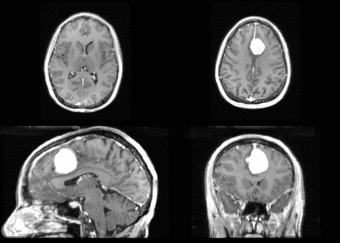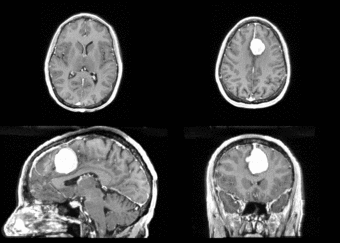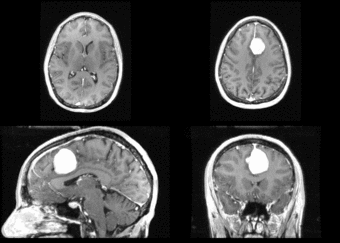
We begin with a tumor growth study. We wish to register the follow up scan to the baseline in order to measure the evolution of the tumor. We perform a rigid registration with six degrees of freedom, three for rotation and | We begin with a tumor growth study. We wish to register the follow up scan to the baseline in order to measure the evolution of the tumor. We perform a rigid registration with six degrees of freedom, three for rotation and | ||
three for translation. Our choice of a rigid transformation is driven by our goal of change assessment, we do not want to introduce scaling during registration. Data taken from the [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseExample1b Registration Case Library]. | three for translation. Our choice of a rigid transformation is driven by our goal of change assessment, we do not want to introduce scaling during registration. Data taken from the [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseExample1b Registration Case Library]. | ||
| + | |||
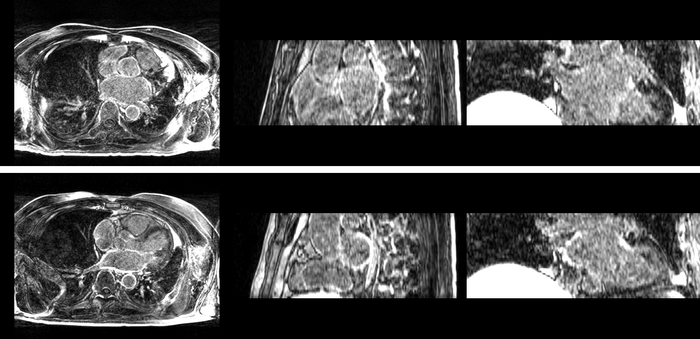
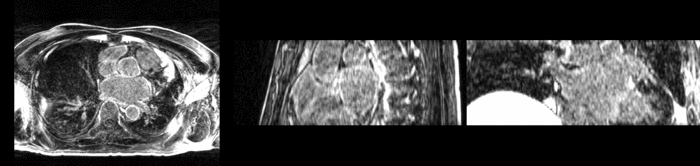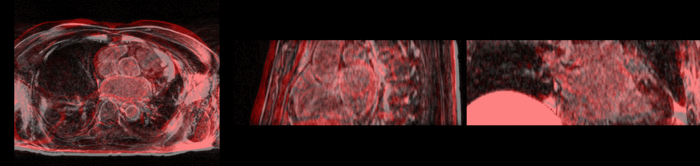
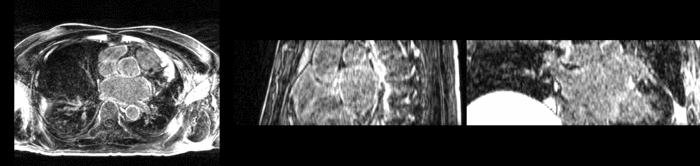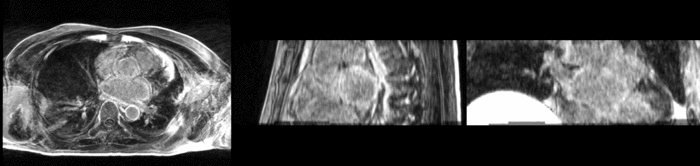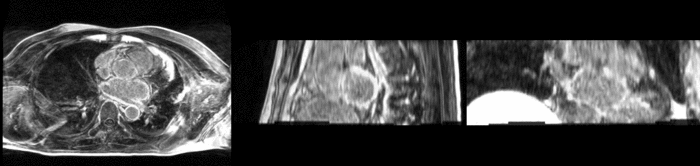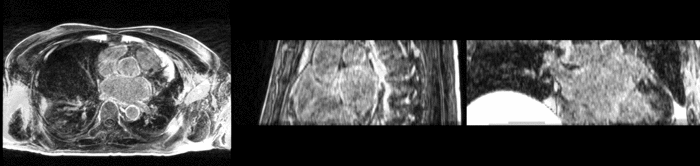
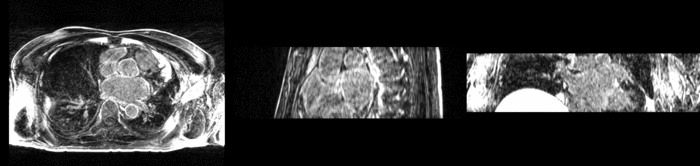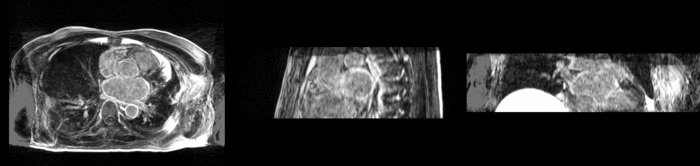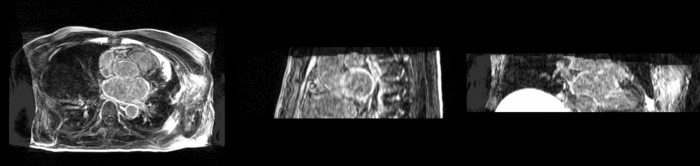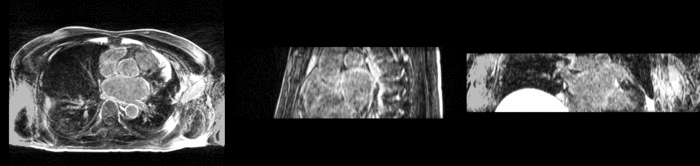
| + | [[Image:All_both.png|thumb|center|700px|Left) First scan. Bottom) Follow up scan.]] | ||
This is an interesting experiment for two reasons. First, tumor change assessment is a common clinical task. As such, there is a need for registration applications to produce high quality results | This is an interesting experiment for two reasons. First, tumor change assessment is a common clinical task. As such, there is a need for registration applications to produce high quality results | ||
| Line 46: | Line 48: | ||
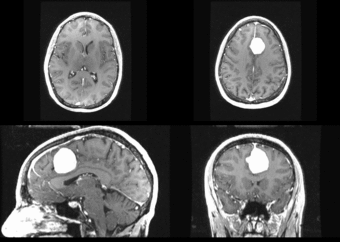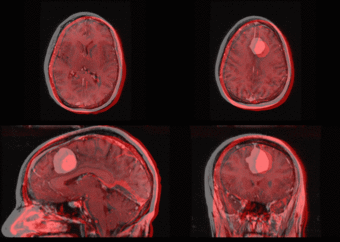
We further observe different bias fields present in the baseline and follow up images. As a result, we choose a similarity metric that is not sensitive to intensity difference, namely mutual information. | We further observe different bias fields present in the baseline and follow up images. As a result, we choose a similarity metric that is not sensitive to intensity difference, namely mutual information. | ||
| − | + | [[Image:tumor_before.gif|thumb|center|340px| Alignment before registration. First scan in gray, follow up scan in red.]] | |
| − | |||
| − | [[Image:tumor_before.gif|thumb|340px| Alignment before registration. First scan in gray, follow up scan in red.]] | ||
[[Image:rview_tumor_reg.gif|thumb|340px| After registration using IRTK.]] | [[Image:rview_tumor_reg.gif|thumb|340px| After registration using IRTK.]] | ||
Revision as of 18:37, 10 May 2010
Home < Projects:RegistrationTestbedContents
Introduction
Image registration is a common step in medical imaging research pipelines. We must establish a common coordinate system across a set of images in order to perform meaningful analysis of the data.
A researcher interested in registering images has many options available. Dozens of algorithms for linear and non-linear registrations have been proposed and are well described in the literature. One possible option for the researcher is to implement the method they feel works best for their specific task. This works well for experts in registration, who are also proficient in software engineering. However, it is unclear how to systematically test registration for correctness. Testing with synthetic examples is possible, but we are ultimately interested in registration of real images. A useful method for testing/validation is comparison to existing registration applications.
Developing a custom registration tool is not a reasonable option for non-experts, for example a doctor performing a clinical study. A user in this case must choose an existing registration application. However, there are many registration packages available, many of which implement the same technique. It is difficult to know which algorithm is well suited for a given registration task, and furthermore, which implementation is best.
Most users fall into the category of non-experts in registration. They will likely use whatever registration package they are currently familiar with or seek the advice of a colleague. At this point, the user is exposed to another layer of difficulty -- parameter selection. It is difficult to select appropriate parameter values without knowledge of how image registration works. A frustrated user might conclude that the registration application they are using doesn't work, simply because they have not found suitable parameters for their data.
In response to the difficulties of image registration, we propose an environment where registration applications can be explored, tested, and compared. This registration testbed provides a common interface to several registration packages, allowing comparison experiments to be performed. A user can explore the impact of parameter settings by running experiments with varying parameter values. We also propose a collection of experiments representative of common registration task, such as multi-modal brain images, for benchmarking and comparison between registration packages.
Related Work
Slicer Registration Case Library
The Slicer Registration Case Library is an effort to establish a collection of registration examples with detailed documentation. The goal is to have a comprehensive collection of examples, so a users can refer to an example similar to their own registration task as a reference, including reasonable parameter settings.
Non-Rigid Image Registration Evaluation Project
The Non-Rigid Image Registration Evaluation Project (NIREP) is an evaluation project led by Gary Christensen at the University of Iowa. They are specifically interested in testing various deformable registration techniques, comparing them to existing registration applications such as SPM2, AIR, and ITK implementations. Furthermore, they propose the development of metrics for measuring the quality of the registration without knowledge of the true solution.
Registration Testbed
Our proposed framework is similar in spirit to that of Batchmake. At the highest level of abstraction, the testbed is a common interface to command line registration applications. It allows for batch processing large experiments for single or multiple registration pipelines. We use the term pipeline to describe a collection of registration applications that are associated with each other. For example, the IRTK pipeline consists of three applications: one for linear registration, one for non-linear registration, and finally an application to produce the output image. While there are three separate applications, they are used together for a single registration task. A pipeline in our testbed consists of one or more registration applications.
At a high level, the testbed has three main components. First, the database represents a collection of definitions of known registration pipelines. This includes knowledge of required and optional input parameters and format of external parameter files used as input. The definitions allow for the implicit enforcement of any rules a given application might have. For example, an application might require a target and source image as the first two arguments, or optional parameters be specified with a "-" or "--". In other words, the database of known definitions helps distance a user from application specific details. This way, a user can more easily generate experiments with one or more registration pipelines without having to know how to use that specific application or consult various documentation. Any command line based registration application can be easily added to the database.
Experiments are stored in an XML format. An experiment file consists of necessary experiment information, such as paths to executables and parameter values. The main testbed application receives an experiment file as input, and using the database, formats the correct command line calls to conduct the experiment. An advanced user can interact directly with the XML to develop experiments. However, a user interface is available to help automate the process.
Examples
Intra-Subject Brain MRI: Axial T1 Tumor Growth Assessment
We begin with a tumor growth study. We wish to register the follow up scan to the baseline in order to measure the evolution of the tumor. We perform a rigid registration with six degrees of freedom, three for rotation and three for translation. Our choice of a rigid transformation is driven by our goal of change assessment, we do not want to introduce scaling during registration. Data taken from the Registration Case Library.
This is an interesting experiment for two reasons. First, tumor change assessment is a common clinical task. As such, there is a need for registration applications to produce high quality results for this type of data. Secondly, the presence of pathology makes registration challenging. If the difference between structures in the two images is considerable, registration algorithms will often produce results of poor quality. This often manifests in poor alignment for rigid registration, or in a large amount of deformation in the area of difference in a non-linear registration.
We further observe different bias fields present in the baseline and follow up images. As a result, we choose a similarity metric that is not sensitive to intensity difference, namely mutual information.