Difference between revisions of "AHM 2007:Slicer3 Developer Feedback"
| Line 52: | Line 52: | ||
===What I would like in Slicer3 base...=== | ===What I would like in Slicer3 base...=== | ||
* More documentation, comments, etc. | * More documentation, comments, etc. | ||
| − | * For CLP tractography in whole brain: ability to seed in RAS, return tracts as models with an attribute (?) saying they're tracts for subsequent conversion to fiberBundleNodes | + | * For CLP tractography in whole brain: ability to seed in RAS, return tracts as models with an attribute (?) saying they're tracts for subsequent conversion to fiberBundleNodes (Should this be CLP or interactive where user can see fibers as seeded?) |
| − | (Should this be CLP or interactive where user can see fibers as seeded?) | + | * Efficient bundle display... |
=EM= | =EM= | ||
Revision as of 18:15, 10 January 2007
Home < AHM 2007:Slicer3 Developer FeedbackBack to AHM_2007#Wednesday.2C_Jan_10_2007:Project_Activities
Contents
Topics for this session:
- Information to the developer community about how these projects have approached developing their code for slicer3 (what's the architecture of their modules)
- Feedback to the base developers about what was easy or hard in the code writing
- What would they like to see in the slicer3 base and/or in VTK, ITK, KKWidgets, etc to make the module development more effective
DTI
DTI Module: Raul San Jose
- The main achivement has been to move slicer from Scalar Volumes to other type of volumetric types: DTI, Stress Tensor, fMRI
- How to integrate new volumes types: DTI support
Core Features
The core features for Diffusion weighted and diffusion tensor images support and visualization have been developed. The main development has been carried out in two fronts:
- MRML library: Volume nodes and display nodes have been created to support the aforementioned volume types.
- Logic layer:
- SlicerSlicerLayer has been extended to support the reslicing of the new node types.
- VolumesLogic: I/O with diffusion data is done through NRRD files using a dedicated vtk library based on teem.
- GUI layer: Volumes interface has been extended to change the look and feel depending on the Volume node selected.
An additional contribution to the core development of DTI infrastructure in the slicer is the addition of different methods for tensor estimation based on teem.
Feedback on Slicer 3
This effort has pushed the code to make easier the addition of new volumes types beyond scalar volumes. The addition of a new volume type requires the addition of code in the following places:
- Creation of MRML nodes and Display MRML nodes
- Addition of a I/O mechanism if not supported by the itk I/O factory and the vtkArchtype wrapping: vtkSlicerVolumesLogic.cxx
- Addition of a reslicing mechanism for the new type and the display logic: vtkSlicerSliceLayerLogic.cxx
- Addition of a GUI class to accomnodate the display options: vtkMyMRMLTypeDisplayGUI.cxx and vtkSlicerVolumesGUI.cxx
Tractography Module: Lauren O'Donnell
Module Architecture
- Improvement from slicer2: Tractography is a real data type in slicer3. Loading, saving, and MRML file I/O will be supported.
- Useful for repeat editing sessions
- Interoperability with other programs
- Tracts saved as "fiber bundle" vtk polydata files with tensors
- There will be several slicer module units to handle tractography. The division will be by Load/Save/Display, Editing (to manually add, select, delete, or to use ROI selection), Seeding, etc.
- Initial work is on the Load/Save/Display and the entire MRML infrastructure.
- MRML details
- Tractography is represented as a vtkMRMLFiberBundleNode. Display parameters are stored in a vtkMRMLFiberBundleDisplayNode, and storage (load/save) handled by vtkMRMLFiberBundleStorageNode. Basic operations of adding new fiber bundles, etc. are in vtkMRMLFiberBundleLogic.
- vtkMRMLFiberBundleDisplayLogic is needed to create "hidden" slicer models that will be rendered in the MRML scene. Otherwise base classes would have to be heavily edited to handle tractography display using the display node.
Feedback on Slicer3
- Most of my feedback is that there is a bit of a learning curve and I spent a few weeks reading over code before doing anything in my module. However Steve and Alex have been very helpful. Specific things that were/are difficult include:
- When adding a new module and a new datatype, it is difficult to find the places in Base code that must be edited.
- It is also difficult to understand how the MRML callback happens, and how to set a new one up. It would be great to have a simple overview on the observers somewhere.
- If a vtkMRMLNode inherits from another one used in slicer (for example mine inherits from the modelNode currently) then the subclass will show up on all of the superclass menus. This is because the vtk "IsA" function is used rather than a direct match. I believe this is feature that is not desired.
- I also have trouble understanding how the MRML file is read in, and how my module can do the "special" things it needs to do (set up logic for display of nodes read in).
- Finally, I would love a tutorial or documentation on the existing interactor functionality in 2D/3D (how do I get mouse clicks in RAS coordinates?, what's the best way to add a picker to the scene?)
What I would like in Slicer3 base...
- More documentation, comments, etc.
- For CLP tractography in whole brain: ability to seed in RAS, return tracts as models with an attribute (?) saying they're tracts for subsequent conversion to fiberBundleNodes (Should this be CLP or interactive where user can see fibers as seeded?)
- Efficient bundle display...
EM
Primary Developers
Yumin Yuan, Sebastien Barre, Brad Davis
Module Description
The following description is from the project wiki page:
The goal of this project is the creation of a module in Slicer 3 integrating the EMSegment algorithm (Pohl et al.), an automatic segmentation algorithm for medical images that previously existed in Slicer 2. As in Slicer 2, the user is able to adjust the algorithm to a variety of imaging protocols as well as anatomical structures and run the segmenter on large data set. However, the configuration of the algorithm is greatly simplified in Slicer 3 as the user is guided by a work flow. The target audience for this module is someone familiar with brain atlases and tissue labels, not a computer scientist.
As of January 1, 2007, the EMSegment module is substantially complete and has been checked into the Slicer3 SVN repository. It was previewed at the December 2006 NAMIC meeting in Clifton Park and will be a demonstration at the NAMIC All-Hands meeting in Salt Lake City on Wednesday 10 January 2007. Future work includes adding advanced and experimental algorithm parameters, improving visualization of parameter settings, and incorporating tissue labels from a controlled vocabulary.
Architecture
The module is implemented as a programmatic Slicer3 module because it requires a large degree of interaction with the user, the data stored in the MRML tree, and the Slicer3 GUI itself. Because the MRML node structure is rather complicated (for example the anatomical tissue hierarchy and a large number of interdependent nodes) the Logic class is solely responsible for maintaining and accessing these nodes. The Logic class provides an API that the GUI code uses to access and modify data. The Logic class also wraps the algorithm code itself.
--Noby 09:25, 10 January 2007 (EST) I am sorry for adding this my personal question but why did you store your parameters in MRML not as attributes of a class? Is this for separating GUI and LOGIC? IGT team (below) is libaries and module, and your inputs from this experience is important for us.
- the parameters for the algorithm are stored in a number of MRML nodes:
- EMSGlobalParametersNode (global algorithm parameters)
- EMSTreeNode (tree structure for hierarchy of anatomical structures)
- EMSTreeParametersNode (algorithm parameters assigned to each tree node)
- EMSAtlasNode (image data and parameters for atlas)
- EMSTargetNode (image data for target images (to be segmented))
- EMSVolumeCollectionNode (essentially a hash map of image volumes, used for atlas and target nodes)
- EMSTemplateNode (collects global parameters and tree structure)
- EMSSegmenterNode (collectes template and output data)
- EMSNode (highest level node, reference to EMSegmenterNode and storage location for module specific data)
- the logic is responsible for
- maintaining and traversing the data in the mrml scene
- providing an interface for the GUI to access the data
- providing an interface for the segmentation algorithm
GUI
The EM GUI includes eight steps implemented with KWWidgets and vtkKWWizardWorkflow framework. Each step is written as a single class, derived from a super class vtkEMSegmentStep. All steps are initialized with parameters from the vtkKWEMSegmentLogic class, and once a user interacts with the UI, the new parameters (if there are) will be passed to the logic class immediately.
There is also a brief description of these steps in the project wiki page:
- 1/8 vtkEMSegmentParametersSetStep: Select parameter set or create new parameters
This can be used as a convenient method to reload all the parameters of a previous EM procedure.
- 2/8 vtkEMSegmentAnatomicalStructureStep: Define a hierarchy of anatomical structures
The anatomical structures are represented with a a tree structure using vtkKWTree, which can be directly manipulated to assign attributes to each anatomical structure, such as name, color, etc.
- 3/8 vtkEMSegmentSpatialPriorsStep: Assign atlases for anatomical structures
Again, the same tree structure is used here to assign atlas to each anatomical structure.
- 4/8 vtkEMSegmentIntensityImagesStep: Choose the set of images that will be segmented
The widget used here, vtkKWListBoxToListBoxSelectionEditor, allows selection of images in different orders, which is actually important in following steps.
- 5/8 vtkEMSegmentIntensityDistributionsStep: Define intensity distribution for each anatomical structure
Same tree structure is used for anatomical structures. This step is where EM module is interacting directly with Slicer3 main application, namely the mouse events from vtkSlicerSliceGUI are processed to get sampling intensities and coordinates.
- 6/8 vtkEMSegmentNodeParametersStep: Specify node-based segmentation parameters
There are many KWWidgets used here (vtkKWTree, vtkKWNotebook, vtkKWMultiColumnList, vtkKWEntryWithScale etc), and given the limited UI space we have , it does take some creativity to arrange everything nicely.
- 7/8 vtkEMSegmentRegistrationParametersStep: Specify atlas-to-target registration parameters
A simple step to assign registration parameters.
- 8/8 vtkEMSegmentRunSegmentationStep: Save work and apply EM Algorithm to segment target images
Again, this step has many gui components.
Feedback
- the community was extremely helpful!
- over all our experience was very positive, but more examples and documentation would be helpful
- used wiki page describing how to build a module---this documentation was helpful
- used GradientAnisotropic module as a template---more examples (and more complicated examples) would be helpful
- we had to write new MRML classes because the slicer2 classes used nested tags and Slicer3 uses references
- could our mrml tree class be generalized for others to use?
- it seems like the creation of most MRML classes could be automated
- it will be very helpful to have a wiki page describing how to observe and process events from main application, such as MRML nodes events, mouse events, progress events etc.
- for such a mid-size project involving several people, we found it would be more productive to develop the whole module in a separate CVS repository (or SVN sandbox), then merge our module with the main Slicer3 repository once it had reached a fair level of stability.
- seperating logic from GUI was a key aspect of developing this module, as it allowed Brad David to focus on the logic, while Yumin and myself (Sebastien) would write the GUI. We started by defining a clean API for the logic and a Powerpoint mockup for the GUI. Brad wrote the corresponding API header and an empty implementation, while we would code the GUI elements and connect them to the logic through the API. Soon after, we added methods to stress-test the API and populate Slicer3 nodes with fake-data so as to test the functionalities of both th elogic and the GUI. Once the piece of the puzzles were in and the most critical bugs were found, we included a real-life dataset for testing.
IGT
PPT file of the presentation at AHM
Noby Hata, Haiying Liu, Simon DiMaio and Raimundo Sierra
Hisotory of our involvement
- Summer 2006: Haiying Liu joined the core development team and translated MRAblation module from the Slicer 2.6. There he
- Learned the architecture of Slicer 3
- Got familiar with KWWidgets
- Fall 2006: Haiying Liu implemented IGT Demo module. There he
- Contributed vtkIGTDemoLogic and vtkIGTDemoGUI classes in the Base
- Added an option for CMake to link the external library OpenTracker
- December 2006: Two additional members joined IGT-Slier3 task force
- Simon DiMaio representing prostate robot project [PI Tempany1R01CA111288]
- Raimundo Sierra representing Neuroendoscope project [PI Jolesz 5U41RR019703]
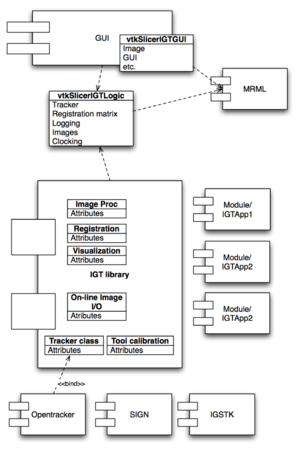
- January 4th, 2007 at 1249 Boylston Office: Implementation strategy for IGT-Slicer has been discussed and preliminary design proposed by Hata and Liu (see the figure at the left)
- Slicer3/Base/GUI/vtkSlicerIGTGUI: cxx class
- Creates GUI components for IGT application modules
- Handles interface update
- Slicer3/Base/Logic/vtkSlicerIGTLogic: cxx class
- Processes shared logics for IGT applications, such as handling communication with tracking server
- Wraps tracker-specific logics
- Slicer3/Libs/IGT: IGT lib (Place holder classes committed to SVN (NH)
- Patient to image registration
- Tool calibration
- Specific tracker logics
- Online image I/O
- Special image processing
- More...
- Slicer3/Modules: IGT applications
- MRAbration
- Neurosurgery
- ProstateBiosy
- More...
- Slicer3/Base/GUI/vtkSlicerIGTGUI: cxx class
Our timeline
- IGT tutorial demo on alpha (done)
- IGT tutorial demo on beta (during AHM week by Liu)
- Neurosurgical navigation with GE Nav (spring to summer)
- Building with IGSTK (during AHM week, by Hata)
- Neurosurgical navigationi with JHU robot (by March)
- Integration to MR/T (fall)
Feedback
- genutiltest.tcl is great!
- Guideline for creating library and modules would be helpful for those joining Slicer community. Template (blank) code may be helpful.
- Don't understand what "NA-MIC sandbox" means...
- It is a bit confusing to request "NA-MIC sandbox" account when you want to create account for "Slicer 3."
- It is nice to have separate account policy for main SVN tree and branches. We want to make our branch relatively open to the IGT community, while the main SVN tree may want to ensure stable release.
- Application-specific customization and streamlining of the GUI will be important for IGT applications. To what extent is this possible?
- Performance bounds are important to understand for IGT applications. Can we talk about determinism and performance as Slicer scales?
- Can we make "lean" Slicer w/o many modules and libraries.
- I found EMsegmentor can be disabled by uncommenting #EMSEG_DEBUG Slicer.cxx, but we don't have an option for that for Tractography, Registrations, etc. We should make this on/off option consistent throughout the Slicer.
- How to specify module dependency
- README.txt to add a module
- What we can do and cannot do with a command line module