Difference between revisions of "2012 Winter Project Week:EndoSeg"
From NAMIC Wiki
| Line 3: | Line 3: | ||
Image:PW-SLC2012.png|[[2012_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2012.png|[[2012_Winter_Project_Week#Projects|Projects List]] | ||
</gallery> | </gallery> | ||
| + | |||
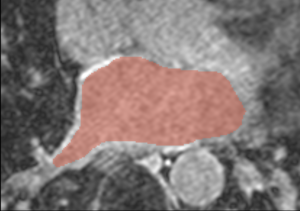
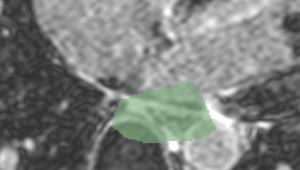
| + | [[File:CARMA_N19_Atlas_LGE.png|thumb|left|Atlas Image]] [[File:CAMRA_N26_LGE.png|thumb|left|Non-atlas Image]] | ||
==Key Investigators== | ==Key Investigators== | ||
Revision as of 22:48, 6 January 2012
Home < 2012 Winter Project Week:EndoSegKey Investigators
Yi Gao, Liang-Jia Zhu, Josh Cates, Alan Morris, Danny Perry, Greg Gardner, Rob MacLeod, Sylvain Bouix, Allen Tannenbaum
Objective
- We are developing methods for segmenting the endocardium from the DE-MRI. The current proposed method is a multi-atlas based registration approach. The user is presented with a weighted average of all registered atlas segmentations. The final LA segmentation is a thresholded volume from this weighted average. Weighting is based on the accuracy of the registration, as determined by a mutual information metric.
- There is a prototype module developed by Yi that is currently in Slicer. We propose to expand and tweak this module to specifications determined by our experience using the algorithm on many real patient datasets.
- We will also discuss posssible collaborative paper ideas based on this work.
Approach, Plan
Approach to be filled here.
Progress
The current segmentation module is in the Slicer extension manager.
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below)
- Slicer Module
- Extension -- commandline