Difference between revisions of "2014 Project Week:Multi-Tissue Stroke Segmentation"
From NAMIC Wiki
| Line 5: | Line 5: | ||
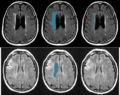
Image:WMH_T1.png| T1 images in stroke dataset. | Image:WMH_T1.png| T1 images in stroke dataset. | ||
Image:WMHseg.png | left: FLAIR images, middle: manual delineation of relevant areas, right: manual WMH segmentation. | Image:WMHseg.png | left: FLAIR images, middle: manual delineation of relevant areas, right: manual WMH segmentation. | ||
| + | Image:Stroke lesion mixture model.jpg | Gaussian mixture model for multimodal intensities. yellow: stroke, blue: normal tissue, red: artifacts, green: WMH. | ||
</gallery> | </gallery> | ||
Latest revision as of 00:58, 10 January 2014
Home < 2014 Project Week:Multi-Tissue Stroke Segmentation
Key Investigators
- Ramesh Sridharan, Adrian Dalca, Polina Binder, Polina Golland, MIT
- Natalia Rost, Jonathan Rosand, MGH
Project Description
Objective
We have developed some methods for segmentation of white matter hyperintensity (WMH) in FLAIR images of stroke patients. We want to extend our framework to do multi-modal segmentation of multiple tissue types (in our case, stroke lesions, white matter hyperintensity, and normal tissue using T1, FLAIR, DWI, and possibly ADC images). This dataset is particularly challenging due to the low resolution (typically 1mm x 1mm x 7mm) and cropped fields of view in the given images.
Approach, Plan
- Identify intensity and shape signatures of different tissue types across images
Progress
- Implemented mixture model for multimodal intensity
- Segmented stroke and white matter hyperintensity, but oversegments: need to find a way to be more specific