Difference between revisions of "PET Tumor Segmentation"
From NAMIC Wiki
| Line 49: | Line 49: | ||
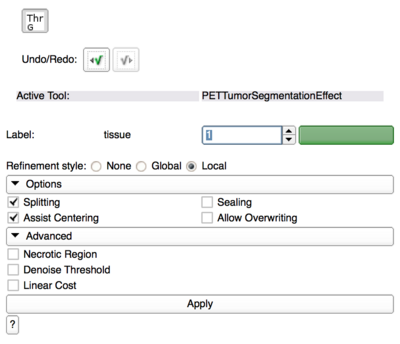
[[File:PETTumorSegmentationEffect_options.png|400px|PET Tumor Segmentation Editor Effect Options]] | [[File:PETTumorSegmentationEffect_options.png|400px|PET Tumor Segmentation Editor Effect Options]] | ||
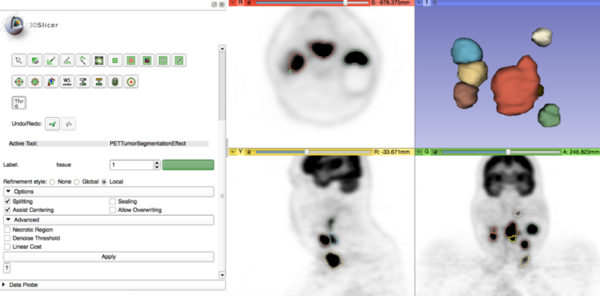
[[File:PETTumorSegmentation_Effect_with_models.png|600px|PET Tumor Segmentation Editor Effect]] | [[File:PETTumorSegmentation_Effect_with_models.png|600px|PET Tumor Segmentation Editor Effect]] | ||
| + | |||
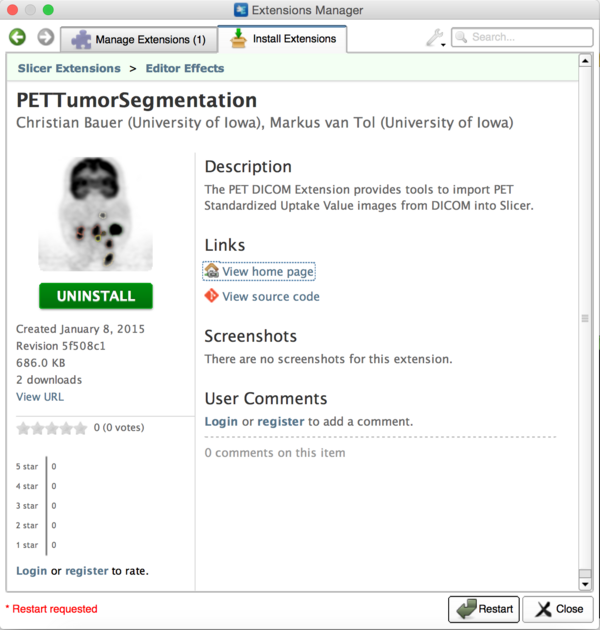
| + | [[File:Extension_Managers_PETTumorSegmentation.png|600px|PET Tumor Segmentation Extension in Extension Manager]] | ||
Revision as of 00:16, 9 January 2015
Home < PET Tumor SegmentationKey Investigators
Christian Bauer (University of Iowa)
Project Description
- Tool webpage: http://www.slicer.org/slicerWiki/index.php/Documentation/Nightly/Modules/PETTumorSegmentationEffect
- Github repository: https://github.com/chribaue/PETTumorSegmentation.git
Objective
- Finalize tool for tumor and lymph node segmentation in PET scans
- Make it publicly available through Slicer Extension Manager
- Segmentation method based on Markus van Tol (2014): A Graph-Based Method for Segmentation of Tumors and Lymph Nodes in Volumetric PET Images, The University of Iowa
Approach, Plan
- Main functionality of the tool has already been developed
- Write self tests
- replace solver with Slicer-license compatible solver
- fine-tune GUI, prepare icon, etc.
- document and clean up source code
- create webpage with documentation and usage tutorial
- get module into Slicer extension management system
- potentially: add functionality to obtain quantitative measurements immediately during segmentation process
Progress
- wrote self tests
- cleaned source code
- switched to new solver and verified correct behavior
- switched to most recent version of 3D Slicer and verified correct behavior
- created webpage with user documentation
- put extension onto github and submitted for integration into Slicer extensions manager