Difference between revisions of "2017 Winter Project Week/MeningiomaSegmentation"
From NAMIC Wiki
(Added examples) |
|||
| Line 8: | Line 8: | ||
==Key Investigators== | ==Key Investigators== | ||
<!-- Add a bulleted list of investigators and their institutions here --> | <!-- Add a bulleted list of investigators and their institutions here --> | ||
| + | * Jakub Kaczmarzyk, MIT | ||
* Satrajit Ghosh, MIT | * Satrajit Ghosh, MIT | ||
* Omar Arnaout, Brigham and Women's Hospital | * Omar Arnaout, Brigham and Women's Hospital | ||
| Line 39: | Line 40: | ||
* Get more data, and potentially train a neural network. | * Get more data, and potentially train a neural network. | ||
|} | |} | ||
| + | |||
| + | |||
| + | ==Examples== | ||
| + | {| class="wikitable" | ||
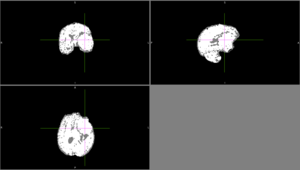
| + | |Sometimes, ANTs failed to remove parts of the skull close to the tumor or wrongly removed part of the brain. | ||
| + | [[File:Case_001_ants_brain_failure.png|thumbnail|ANTs brain extraction failure]] | ||
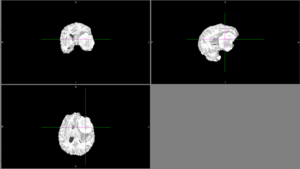
| + | But other times, ANTs extracted the brain very well. | ||
| + | [[File:Case_052_ants_brain.png|thumbnail|Ants brain extraction success]] | ||
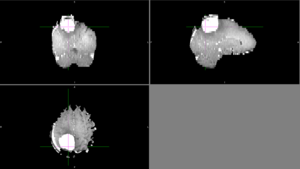
| + | Automatic segmentation with ANTs usually classified as CSF or it classified different parts of the tumor as different types of tissue. | ||
| + | [[File:Case_052_ants_brain_seg.png|thumbnail|Brain segmentation with ANTs]] | ||
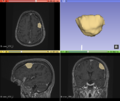
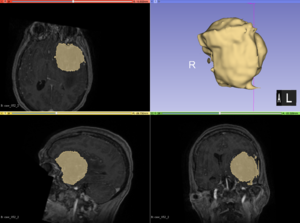
| + | Semi-automatic segmentation with Slicer was relatively successful. Sometimes the segmentation would bleed outside of the tumor into voxels with similar intensities. | ||
| + | [[File:Case_052_2_slicer_seg.png|thumbnail|Semi-automatic segmentation with Slicer]] | ||
| + | | | ||
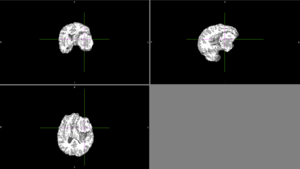
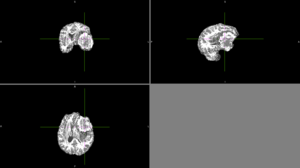
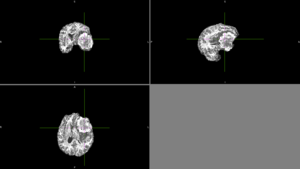
| + | We tried FSL's FAST with different numbers of classes. None of these methods could identify the entire tumor mass as one type of tissue in this scan. | ||
| + | [[File:Case_052_fast_3classes.png|thumbnail|FAST brain segmentation with 3 classes]] | ||
| + | [[File:Case_052_fast_4classes.png|thumbnail|FAST brain segmentation with 4 classes]] | ||
| + | [[File:Case_052_fast_5classes.png|thumbnail|FAST brain segmentation with 5 classes]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
==Background and References== | ==Background and References== | ||
<!-- Use this space for information that may help people better understand your project, like links to papers, source code, or data --> | <!-- Use this space for information that may help people better understand your project, like links to papers, source code, or data --> | ||
MR images of meningiomas that will be used in this project are available at [http://openneu.ro/metasearch/ OpenNeu.ro]. | MR images of meningiomas that will be used in this project are available at [http://openneu.ro/metasearch/ OpenNeu.ro]. | ||
Revision as of 21:02, 13 January 2017
Home < 2017 Winter Project Week < MeningiomaSegmentationKey Investigators
- Jakub Kaczmarzyk, MIT
- Satrajit Ghosh, MIT
- Omar Arnaout, Brigham and Women's Hospital
Project Description
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
Progress
Next steps
|
Examples
| Sometimes, ANTs failed to remove parts of the skull close to the tumor or wrongly removed part of the brain.
But other times, ANTs extracted the brain very well. Automatic segmentation with ANTs usually classified as CSF or it classified different parts of the tumor as different types of tissue. Semi-automatic segmentation with Slicer was relatively successful. Sometimes the segmentation would bleed outside of the tumor into voxels with similar intensities. |
We tried FSL's FAST with different numbers of classes. None of these methods could identify the entire tumor mass as one type of tissue in this scan. |
Background and References
MR images of meningiomas that will be used in this project are available at OpenNeu.ro.