Difference between revisions of "Collaboration/WFU/NonHuman Primate Neuroimaging"
From NAMIC Wiki
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:ProjectWeek-2007.png|thumb|320px|Return to [[2007_Programming/Project_Week_MIT|Project Week Main Page]] ]] | |[[Image:ProjectWeek-2007.png|thumb|320px|Return to [[2007_Programming/Project_Week_MIT|Project Week Main Page]] ]] | ||
| − | |||
|} | |} | ||
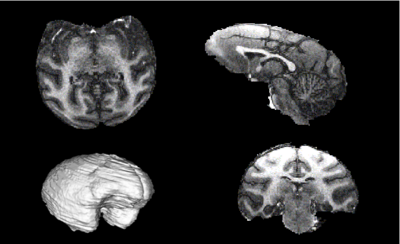
| + | Before: [[Image:Nhpbrainexample.png|400px]] | ||
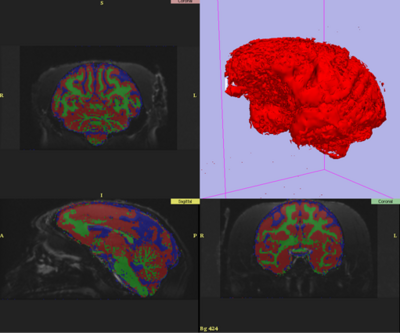
| + | After: [[Image:ProgWeek07NHPResult1.png|400px]] | ||
__NOTOC__ | __NOTOC__ | ||
| Line 44: | Line 45: | ||
During the project week: | During the project week: | ||
| − | * Nonlinear registration of atlas priors completed | + | * Nonlinear registration of atlas priors completed using Slicer |
| − | * Full segmentation model accommodating severe coil inhomogeneity completed | + | * Full EM segmentation model accommodating severe coil inhomogeneity completed |
| − | * | + | * ITK-based utility to spatially blend prior maps written |
| + | * EMSegmenter parameters partially optimized for a single subject | ||
* Scripts prepared to run the full study | * Scripts prepared to run the full study | ||
</div> | </div> | ||
| Line 53: | Line 55: | ||
</div> | </div> | ||
| − | |||
| − | |||
Revision as of 16:08, 28 June 2007
Home < Collaboration < WFU < NonHuman Primate Neuroimaging Return to Project Week Main Page |
This project is part of an NIH-NIAAA funded "Collaborations with National Centers for Biomedical Computing" grant,
R01-AA016748-01 "Measuring Alcohol and Stress Interactions with Structural and Perfusion MRI" (Jim Daunais PI).
Key Investigators
- VT/WFU: Chris Wyatt
- BWH: Kilian Pohl
Objective
The objectives of this project are to:
- Optimize the EM Brain Classifier in Slicer on MRI images of rhesus macaques
- Develop a custom GM/WM/CSF atlas, bootstrapping from one provided by Martin Styner
- Integrate a sub-cortical atlas
- Segment data from N subjects in the alcohol naive and post-induction state.
- Compare GM/WM volume and ratios between the two states
- Compare hippocampus, putamen, and caudate volumes between states
Approach, Plan
We will use the current implementation of the EM Brain classifier in slicer. Using a manual segmentation as an estimate of ground truth in a single subject, we will optimize the EM segmenter parameters over the classification error and running time. The study will be run via VPN on a Sungrid at VT.
Progress
Prior to the project week:
- Study data on 8 subjects at two time points (alcohol naive and post-induction) organized
- Study data has been skull stripped, ECC masks created, and GM/WM/CSF priors registered to each subject. The ECC masks may need some hand editing and, as a result, the atlas-priors to subject registration is poor in several subjects.
During the project week:
- Nonlinear registration of atlas priors completed using Slicer
- Full EM segmentation model accommodating severe coil inhomogeneity completed
- ITK-based utility to spatially blend prior maps written
- EMSegmenter parameters partially optimized for a single subject
- Scripts prepared to run the full study
References
- K. M. Pohl, J. Fisher, W.E.L. Grimson, R. Kikinis, and W.M. Wells. A Bayesian Model for Joint Segmentation and Registration. NeuroImage, 31(1), pp. 228-239, 2006.
- E. V. Sullivan, H. J. Sable, W. N Strother, D. P. Friedman, A. Davenport, H. Tillman-Smith, R. A. Kraft, C. L. Wyatt, K. T. Szeliga, N. C. Buchheimer, J. B. Daunais, E. Adalsteinsson, A. Pfefferbaum, K. A Grant, Neuroimaging of Rodent and Primate Models of Alcoholism: Initial Reports from the Integrative Neuroscience Initiative on Alcoholism, Alcoholism: Clinical and Experimental Research Vol. 29 No. 2 pgs 287-294 Feb 2005.