Difference between revisions of "Projects:fMRIClustering"
(New page: Back to NA-MIC_Collaborations, MIT Algorithms = fMRI Clustering = In this work we propose to simultaneously estimate the optimal representati...) |
|||
| Line 3: | Line 3: | ||
= fMRI Clustering = | = fMRI Clustering = | ||
| − | In this | + | In the classical functional connectivity analysis, networks of interest are |
| + | defined based on correlation with the mean time course of a user-selected | ||
| + | `seed' region. Further, the user has to also specify a subject-specific threshold at which correlation | ||
| + | values are deemed significant. In this project, we simultaneously estimate the optimal | ||
representative time courses that summarize the fMRI data well and | representative time courses that summarize the fMRI data well and | ||
the partition of the volume into a set of disjoint regions that are best | the partition of the volume into a set of disjoint regions that are best | ||
| − | explained by these representative time courses. | + | explained by these representative time courses. This approach to functional connectivity analysis offers two |
advantages. First, is removes the sensitivity of the analysis to the details | advantages. First, is removes the sensitivity of the analysis to the details | ||
of the seed selection. Second, it substantially simplifies group analysis | of the seed selection. Second, it substantially simplifies group analysis | ||
| − | by eliminating the need for | + | by eliminating the need for the subject-specific threshold. Our experimental results indicate that |
| − | |||
| − | |||
| − | |||
the functional segmentation provides a robust, anatomically meaningful | the functional segmentation provides a robust, anatomically meaningful | ||
and consistent model for functional connectivity in fMRI. | and consistent model for functional connectivity in fMRI. | ||
| + | |||
| + | Currently, we are investigating the application of such clustering algorithms in detection of functional connectivity in high-level | ||
= Description = | = Description = | ||
| − | '' | + | ''Generative Model for Functional Connectivity'' |
| + | |||
| + | |||
| − | + | This unsupervised technique generalizes | |
| + | connectivity analysis to situations where candidate seeds are diffi�cult to | ||
| + | identify reliably or are unknown. | ||
| − | |||
| − | |||
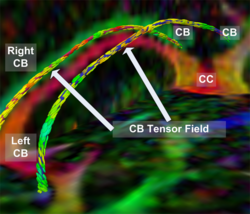
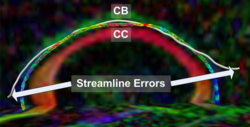
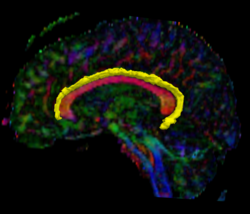
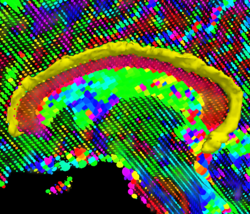
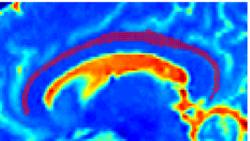
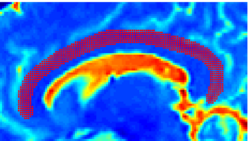
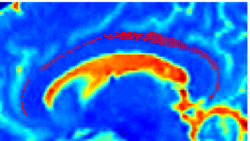
Recently, we have applied this method to the cingulum bundle, as shown in the following images: | Recently, we have applied this method to the cingulum bundle, as shown in the following images: | ||
| Line 39: | Line 43: | ||
''Fiber Bundle Segmentation'' | ''Fiber Bundle Segmentation'' | ||
| − | + | ||
{| | {| | ||
| Line 62: | Line 66: | ||
|} | |} | ||
| − | '' | + | ''Experimental Result'' |
| − | We | + | We used data from 7 subjects with a diverse set of visual experiments including localizer, morphing, rest, internal tasks, and movie. The functional scans were pre-processed for motion artifacts, manually aligned into the Talairach coordinate system, detrended (removing linear trends in the |
| + | baseline activation) and smoothed (8mm kernel). | ||
| − | '' | + | ''Project Status'' |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
*Working 3D implementation in Matlab using the C-based Mex functions. | *Working 3D implementation in Matlab using the C-based Mex functions. | ||
| − | *Currently porting to ITK. | + | *Currently porting to ITK. (last updated 18/Apr/2007) |
= Key Investigators = | = Key Investigators = | ||
| − | * | + | * MIT: Danial Lashkari, Polina Golland, Nancy Kanwisher |
| − | |||
= Publications = | = Publications = | ||
| Line 89: | Line 84: | ||
''In print'' | ''In print'' | ||
| − | + | * P. Golland, Y. Golland, R. Malach. Detection of Spatial Activation Patterns As Unsupervised Segmentation of fMRI Data. In Proceedings of MICCAI: International Conference on Medical Image Computing and Computer Assisted Intervention, 110-118, 2007. | |
''In press'' | ''In press'' | ||
| − | * | + | * D. Lashkari, P. Golland. Convex Clustering with Exemplar-Based Models. In NIPS: Advances in Neural Information Processing Systems, 2007. |
| − | |||
| − | |||
| − | |||
= Links = | = Links = | ||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 20:33, 9 November 2007
Home < Projects:fMRIClusteringBack to NA-MIC_Collaborations, MIT Algorithms
fMRI Clustering
In the classical functional connectivity analysis, networks of interest are defined based on correlation with the mean time course of a user-selected `seed' region. Further, the user has to also specify a subject-specific threshold at which correlation values are deemed significant. In this project, we simultaneously estimate the optimal representative time courses that summarize the fMRI data well and the partition of the volume into a set of disjoint regions that are best explained by these representative time courses. This approach to functional connectivity analysis offers two advantages. First, is removes the sensitivity of the analysis to the details of the seed selection. Second, it substantially simplifies group analysis by eliminating the need for the subject-specific threshold. Our experimental results indicate that the functional segmentation provides a robust, anatomically meaningful and consistent model for functional connectivity in fMRI.
Currently, we are investigating the application of such clustering algorithms in detection of functional connectivity in high-level
Description
Generative Model for Functional Connectivity
This unsupervised technique generalizes connectivity analysis to situations where candidate seeds are diffi�cult to identify reliably or are unknown.
Recently, we have applied this method to the cingulum bundle, as shown in the following images:
Fiber Bundle Segmentation
Experimental Result
We used data from 7 subjects with a diverse set of visual experiments including localizer, morphing, rest, internal tasks, and movie. The functional scans were pre-processed for motion artifacts, manually aligned into the Talairach coordinate system, detrended (removing linear trends in the baseline activation) and smoothed (8mm kernel).
Project Status
- Working 3D implementation in Matlab using the C-based Mex functions.
- Currently porting to ITK. (last updated 18/Apr/2007)
Key Investigators
- MIT: Danial Lashkari, Polina Golland, Nancy Kanwisher
Publications
In print
- P. Golland, Y. Golland, R. Malach. Detection of Spatial Activation Patterns As Unsupervised Segmentation of fMRI Data. In Proceedings of MICCAI: International Conference on Medical Image Computing and Computer Assisted Intervention, 110-118, 2007.
In press
- D. Lashkari, P. Golland. Convex Clustering with Exemplar-Based Models. In NIPS: Advances in Neural Information Processing Systems, 2007.