Difference between revisions of "Projects:KPCA LLE KLLE ShapeAnalysis"
| Line 23: | Line 23: | ||
''In Print'' | ''In Print'' | ||
* [http://www.na-mic.org/pages/Special:Publications?text=Projects%3AKPCA+LLE+KLLE+ShapeAnalysis&submit=Search&words=all&title=checked&keywords=checked&authors=checked&abstract=checked&sponsors=checked&searchbytag=checked| NA-MIC Publications Database] | * [http://www.na-mic.org/pages/Special:Publications?text=Projects%3AKPCA+LLE+KLLE+ShapeAnalysis&submit=Search&words=all&title=checked&keywords=checked&authors=checked&abstract=checked&sponsors=checked&searchbytag=checked| NA-MIC Publications Database] | ||
| + | |||
| + | [[Category: Shape Analysis]] | ||
Revision as of 17:46, 1 April 2008
Home < Projects:KPCA LLE KLLE ShapeAnalysisBack to Georgia Tech Algorithms
KPCA LLE KLLE Shape Analysis
Our Objective is to compare various shape representation techniques like linear PCA (LPCA), kernel PCA (KPCA), locally linear embedding (LLE) and kernel locally linear embedding (KLLE).
Description
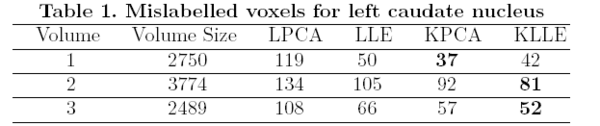
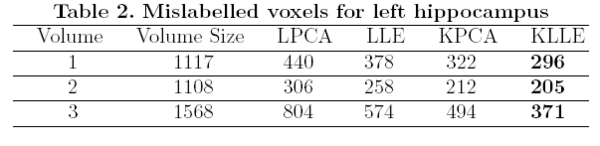
The surfaces are represented as the zero level set of a signed distance function and shape learning is performed on the embeddings of these shapes. We carry out some experiments to see how well each of these methods can represent a shape, given the training set. We tested the performance of these methods on shapes of left caudate nucleus and left hippocampus. The training set of left caudate nucleus consisted of 26 data sets and the test set contained 3 volumes. Error between a particular shape representation and ground truth was calculated by computing the number of mislabeled voxels using each of the methods. Figure 1 gives the error using each of the methods. Similar tests were done on a training set of 20 hippocampus data with 3 test volumes. Figure 2 gives the error table for each of the methods [1].
Key Investigators
- Georgia Tech Algorithms: Yogesh Rathi, Samuel Dambreville, Allen Tannenbaum
Publications
In Print