Difference between revisions of "Projects:DTIPopulationAnalysis"
(Merge the quantitative tract analysis project) |
(Reformat to combine atlas building and quantitative tract analysis projects) |
||
| Line 17: | Line 17: | ||
|} | |} | ||
| − | |||
Our registration procedure is based on a scalar feature image which is sensitive to sheet like structures. We have observed that the major fiber bundles of interest occur as sheet or tube like manifolds in the FA image of the brain. As a feature image we use the maximum eigenvalue of the hessian of the FA image. Images are initially aligned using an affine registration and then deformed to a common coordinate system using the unbiased atlas-building procedure of Joshi et al. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15501084&dopt=Citation]. The deformation fields produced by the registration process are applied to the tensors fields using appropriate methods for reorienting and interpolating tensors. The transformed images are averaged in the atlas space to produce a DTI atlas. | Our registration procedure is based on a scalar feature image which is sensitive to sheet like structures. We have observed that the major fiber bundles of interest occur as sheet or tube like manifolds in the FA image of the brain. As a feature image we use the maximum eigenvalue of the hessian of the FA image. Images are initially aligned using an affine registration and then deformed to a common coordinate system using the unbiased atlas-building procedure of Joshi et al. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15501084&dopt=Citation]. The deformation fields produced by the registration process are applied to the tensors fields using appropriate methods for reorienting and interpolating tensors. The transformed images are averaged in the atlas space to produce a DTI atlas. | ||
| Line 26: | Line 25: | ||
We have begun to apply the DTI atlas building procedure to data provided by the [[DBP:Harvard|PNL]]. A combined set of DTI scans from control and Schizophrenic subjects were aligned using the procedure described above. In the atlas space the SZ and CNTL groups are processed to produce voxel-wise statistics for each group. The figure below shows colored FA and mean diffusivity slices for both the CNTL and SZ group. Preliminary work is now being done on region of interest (ROI) hypothesis testing between the two populations. | We have begun to apply the DTI atlas building procedure to data provided by the [[DBP:Harvard|PNL]]. A combined set of DTI scans from control and Schizophrenic subjects were aligned using the procedure described above. In the atlas space the SZ and CNTL groups are processed to produce voxel-wise statistics for each group. The figure below shows colored FA and mean diffusivity slices for both the CNTL and SZ group. Preliminary work is now being done on region of interest (ROI) hypothesis testing between the two populations. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{| | {| | ||
| Line 44: | Line 35: | ||
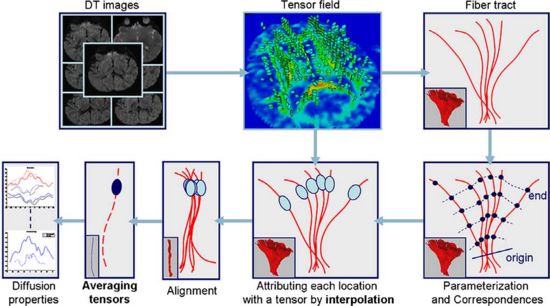
This project proposes a framework for quantitative analysis of DTI data. The framework uses the full tensor information for statistical analysis using the affine-invariant Riemannian metric for defining operations such as interpolation and averaging on tensors. Furtheremore, the results of tractography are used to provide a reference coordinate system the respresents the underlying structure of fiber bundles. The tract modeling framework includes a model both of the geometry of the fiber bundle and of the diffusion properties along the bundle. A new anisotropy measure called geodesic anisotropy (GA) is also included in the framework. | This project proposes a framework for quantitative analysis of DTI data. The framework uses the full tensor information for statistical analysis using the affine-invariant Riemannian metric for defining operations such as interpolation and averaging on tensors. Furtheremore, the results of tractography are used to provide a reference coordinate system the respresents the underlying structure of fiber bundles. The tract modeling framework includes a model both of the geometry of the fiber bundle and of the diffusion properties along the bundle. A new anisotropy measure called geodesic anisotropy (GA) is also included in the framework. | ||
| − | |||
| − | |||
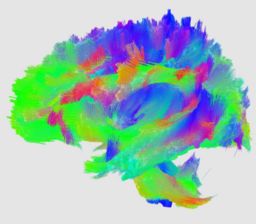
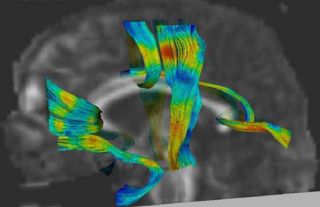
[[image:corouge_dti_statistics.jpg|thumb|320px|Fiber tracts colored with FA attributes]] | [[image:corouge_dti_statistics.jpg|thumb|320px|Fiber tracts colored with FA attributes]] | ||
[[image:corouge-tract-analysis-flowchart.jpg|550px]] | [[image:corouge-tract-analysis-flowchart.jpg|550px]] | ||
| + | |||
| + | = Group Analysis of tract properties = | ||
| + | We have combined the procedure for atlas building of DTI with the tract analysis to provide a method for tract specific statistical analysis. | ||
= Publications = | = Publications = | ||
Revision as of 19:58, 25 August 2008
Home < Projects:DTIPopulationAnalysisBack to NA-MIC Collaborations, Utah2 Algorithms
Contents
DTI ATlas Building
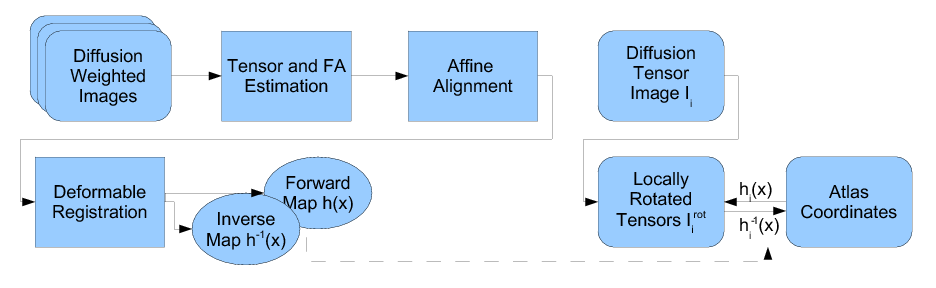
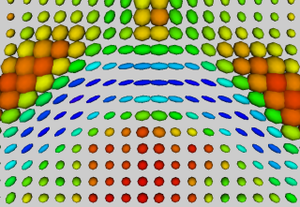
Our methodology for population analysis of DT-MRI is based on unbiased non-rigid registration of a population to a common coordinate system. The registration jointly produces an average DTI atlas, which is unbiased with respect to the choice of a template image, along with diffeomorphic correspondence between each image. The registration image match metric uses a feature detector for thin fiber structures of white matter, and interpolation and averaging of diffusion tensors use the Riemannian symmetric space framework. The anatomically significant correspondence provides a basis for comparison of tensor features and fiber tract geometry in clinical studies.
Our registration procedure is based on a scalar feature image which is sensitive to sheet like structures. We have observed that the major fiber bundles of interest occur as sheet or tube like manifolds in the FA image of the brain. As a feature image we use the maximum eigenvalue of the hessian of the FA image. Images are initially aligned using an affine registration and then deformed to a common coordinate system using the unbiased atlas-building procedure of Joshi et al. [1]. The deformation fields produced by the registration process are applied to the tensors fields using appropriate methods for reorienting and interpolating tensors. The transformed images are averaged in the atlas space to produce a DTI atlas.
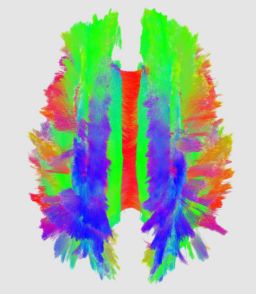
An initial test was performed by using the procedure on a set of images of healthy subject at age one year. The results of the tensor averaging are shown on the right. Tractography was also performed on the mean atlas image as shown.
Collaboration with PNL
We have begun to apply the DTI atlas building procedure to data provided by the PNL. A combined set of DTI scans from control and Schizophrenic subjects were aligned using the procedure described above. In the atlas space the SZ and CNTL groups are processed to produce voxel-wise statistics for each group. The figure below shows colored FA and mean diffusivity slices for both the CNTL and SZ group. Preliminary work is now being done on region of interest (ROI) hypothesis testing between the two populations.
DTI Quantitative Tract Analysis
This project proposes a framework for quantitative analysis of DTI data. The framework uses the full tensor information for statistical analysis using the affine-invariant Riemannian metric for defining operations such as interpolation and averaging on tensors. Furtheremore, the results of tractography are used to provide a reference coordinate system the respresents the underlying structure of fiber bundles. The tract modeling framework includes a model both of the geometry of the fiber bundle and of the diffusion properties along the bundle. A new anisotropy measure called geodesic anisotropy (GA) is also included in the framework.
Group Analysis of tract properties
We have combined the procedure for atlas building of DTI with the tract analysis to provide a method for tract specific statistical analysis.
Publications
Key Investigators
Utah Algorithms: Casey Goodlett, Isabelle Corouge, P. Thomas Fletcher, Guido Gerig
Software
- Algorithms written in ITK. GUI of prototype software written in QT (FiberViewer software). Prototype software tested in clinical studies at UNC. Validation tests with repeated DTI of same subject (6 cases). FiberTracking download
- Additionally available: ITK compatible fibertracking prototype tool FiberTracking to be used to study overlap/dissimilarity with other tools already available to NA-MIC: Functionality: reads raw DW-MRI data (6 direction Basser scheme), fiber tracking based on user-selected source and regions (S. Mori scheme), display of fibertracts and volumetric data, output: sets of streamlines in ITK polyline format attributedwith DTI properties and display parameteres (radiusof tubes, local color, etc.). FiberViewer download
- Command line tools for DTI processing available from UNC NeuroLib