Difference between revisions of "Stanford SIMBIOS: Whole Body Segmentation for Simulation"
| Line 1: | Line 1: | ||
| − | [[Image: | + | {| |
| − | [[Image:ct.jpg]] | + | |[[Image:NAMIC-SLC.jpg|thumb|320px|Return to [[2009_Winter_Project_Week|Project Week Main Page]] ]] |
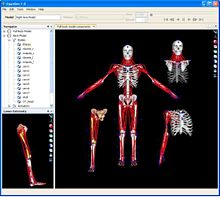
| + | |[[Image:ct.jpg|thumb|190px|Example whole-body CT dataset from WashU displayed in 3D Slicer.]] | ||
| + | |[[Image:sim.jpg|thumb|220px|Articulated musculoskeletal models.]] | ||
| + | |} | ||
===Key Investigators=== | ===Key Investigators=== | ||
* Stanford: Harish Doddi, Saikat Pal, and Scott Delp | * Stanford: Harish Doddi, Saikat Pal, and Scott Delp | ||
Revision as of 21:56, 15 December 2008
Home < Stanford SIMBIOS: Whole Body Segmentation for Simulation Return to Project Week Main Page |
Key Investigators
- Stanford: Harish Doddi, Saikat Pal, and Scott Delp
- WashU: Daniel Marcus
- Harvard: ??
Tutorial
Objective
The aim of this project is to develop an automatic/semi-automatic methodology to convert whole body imaging datasets into three-dimensional models for neuromuscular biomechanics and finite element simulations. Initially, we will investigate the existing capabilities in EMSegmenter software to automatically segment the knee joint.
Approach, Plan
Investigate the existing functionality of EMSegementer to extract whole body models from CT and MR datasets. Initial efforts will be focused on developing atlases of specific joints (e.g. the knee) and evaluating EMSegmenter algorithms. The plan is to have imported MRI knee geometries in EMSegmenter and create an average atlas before the project week. During the project week, the EMSegmenter algorithm will be tested on a specific subject geometry.
Progress