Difference between revisions of "Slicer3.2:Training"
Hjbockholt (talk | contribs) |
Hjbockholt (talk | contribs) |
||
| Line 71: | Line 71: | ||
| style="background:#D1FFF9; color:black"| [[media:Slicer3Training_WhiteMatterLesions_v2.2.1.pdf|Detecting white matter lesions in lupus]] <br> | | style="background:#D1FFF9; color:black"| [[media:Slicer3Training_WhiteMatterLesions_v2.2.1.pdf|Detecting white matter lesions in lupus]] <br> | ||
This tutorial teaches how to use an automated, multi-level method to segment white matter brain lesions in lupus. | This tutorial teaches how to use an automated, multi-level method to segment white matter brain lesions in lupus. | ||
| − | | style="background:#D1FFF9; color:black"|[[Media:LesionSegmentationTutorialData.tgz| | + | | style="background:#D1FFF9; color:black"|Lupus Tutorial Data[[Media:LesionSegmentationTutorialData.tgz|(.tgz)]][[media:LesionSegmentationTutorialData.zip| (.zip)]]<br> |
| style="background:#C3D1C3; color:black" align="Center"| [[Image:3dLupusLesionVolume.png|200px|Lesion Segmentation]] | | style="background:#C3D1C3; color:black" align="Center"| [[Image:3dLupusLesionVolume.png|200px|Lesion Segmentation]] | ||
|- | |- | ||
Revision as of 18:07, 25 June 2009
Home < Slicer3.2:TrainingWelcome to the 3D Slicer3.2 User Training 101
- The information on this page applies to 3D Slicer version 3.2.
- If you are looking for materials about Slicer version 3.4, please click here
- If you are looking for materials about Slicer version 2, please visit the Slicer2 101 page.
|
Slicer Logo | |
Software Installation
The Slicer download page contains links for downloading the different versions of Slicer 3.
Training Compendium
| Category | Tutorial | Sample Data | Image |
| 1.1 | Data Loading and 3D Visualization in Slicer3 | SlicerSampleVisualization.tar.gz SlicerSampleVisualization.zip |
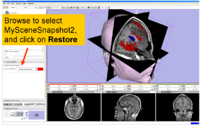
|
| 1.2 | Manual segmentation with 3D Slicer | 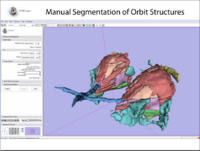
| |
| 1.3 | EM Segmentation Course Background Materials: EMSegmenter History |
AutomaticSegmentation.tar.gz
|

|
| 1.4 | Affine and Deformable Registration in Slicer 3 | SlicerSampleRegistration.tgz |
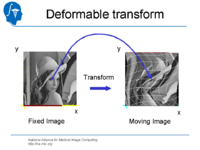
|
| 1.5 | Processing of Diffusion Weighted Imaging and Diffusion Tensor Imaging data in Slicer3 Background Materials:DT-MRI module. |
Slicer3DiffusionTutorialData.zip |
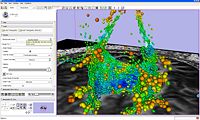
|
| 1.6 | Detecting subtle change in pathology This tutorial consists of two MR scans of a patient with meningioma. |
ChangeTracker-Tutorial-Data.zip | 
|
| 1.7 | Detecting white matter lesions in lupus This tutorial teaches how to use an automated, multi-level method to segment white matter brain lesions in lupus. |
Lupus Tutorial Data(.tgz) (.zip) |
|
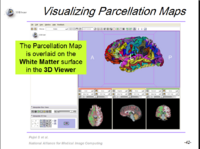
| 1.8 | FreeSurfer Course This tutorial teaches how to visualize FreeSurfer segmentations and parcellation maps of the brain. |
FreeSurfer Tutorial Data FreeSurferTutorialData.zip |
|
| 2.1 | Programming into Slicer3
This tutorial is intended for engineers and scientists who want to integrate stand-alone programs outside of the Slicer3 source tree. |
HelloWorld_Plugin.zip | 
|
| 2.2 | Image Guided Therapy Planning Tutorial This tutorial takes the trainee through a complete workup for neurosurgical patient specific mapping.See pages 58 to 80 of this tutorial on using the "Simple region growing" segmentation module.
|
NeurosurgicalPlanningTutorialData.zip | 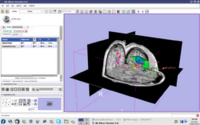
|
| 3.1 | Slicer3 as a research tool for image guided therapy research (IGT)
This tutorial is intended for engineers and scientists who want to use Slicer 3 for IGT research. |

| |
| 3.2 | SPL-PNL Brain Atlas Tutorial
This three-dimensional brain atlas dataset, derived from a volumetric, whole-head MRI scan, contains the original MRI-scan, a complete set of label maps, three-dimensional reconstructions (200+ structures) and a tutorial. |
SPL-PNL Brain Atlas | 
|
| 3.3 | SPL Abdominal Atlas Tutorial
This three-dimensional abdominal atlas was derived from a computed tomography (CT) scan, using semi-automated image segmentation and three-dimensional reconstruction techniques. The dataset contains the original CT scan, detailed label maps, three-dimensional reconstructions and a tutorial.
|
SPL Abdominal Atlas | 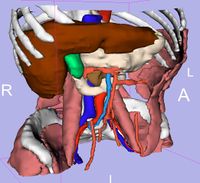
|
| 3.4 | Using XNAT Desktop and Slicer3 for remote data handling
This tutorial is intended for researchers who wish to upload local datasets to a remote XNAT repository like XNAT Central and retrieve catalog files that can be opened inside Slicer. |
| |
| 3.5 | Harnessing Grid Computational Resources and integration with Slicer through the Grid Wizard Enterprise system
These tutorials are intended for researchers who wish to utilize distributed computational resources. |
Grid Wizard Enterprise | 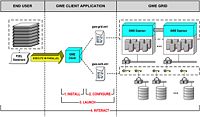 Featured Study Case (YouTube) Featured Study Case (YouTube)
|
- Category 1 = Basic functionality
- Category 2 = Advanced functionality
- Category 3 = Specialized application packages
Additional Materials
For a variety of data sets for downloading, check the following link.
Back to Training:Main
