Difference between revisions of "Projects:LatentAtlasSegmentation"
| Line 62: | Line 62: | ||
Joint Segmentation using Patient specific Latent Anatomy Model, MICCAI workshop for Probabilistic Models on Medical Image Analysis (PMMIA) 2009 | Joint Segmentation using Patient specific Latent Anatomy Model, MICCAI workshop for Probabilistic Models on Medical Image Analysis (PMMIA) 2009 | ||
| − | = Key Investigators | + | = Key Investigators |
| − | + | * MIT, CSAIL [[http://people.csail.mit.edu/tammy/ | Tammy Riklin Raviv]], Polina Golland | |
| − | * MIT [[http://people.csail.mit.edu/tammy/ | Tammy Riklin Raviv]], MGH | + | * Harvard Medical School, MGH | Koen Van Leemput |
| + | * Harvard Medical School, BWH | William M. Wells, Ron Kikinis, Martha Shenton, Sylvain Bouix | ||
Revision as of 19:25, 10 September 2009
Home < Projects:LatentAtlasSegmentationBack to NA-MIC Collaborations, MIT Algorithms,
Joint Segmentation of Image Ensembles via Latent Atlases
Spatial priors, such as probabilistic atlases, play an important role in MRI segmentation. The atlases are typically generated by averaging manual labels of aligned brain regions across different subjects. However, the availability of comprehensive, reliable and suitable manual segmentations is limited. We therefore propose a joint segmentation of corresponding, aligned structures in the entire population that does not require a probability atlas. Instead, a latent atlas, initialized by a single manual segmentation, is inferred from the evolving segmentations of the ensemble. The proposed method is based on probabilistic principles but is solved using partial differential equations (PDEs) and energy minimization criteria. We evaluate the method by segmenting 50 brain MR volumes. Segmentation accuracy for cortical and subcortical structures approaches the quality of state-of-the-art atlas-based segmentation results, suggesting that the latent atlas method is a reasonable alternative when existing atlases are not compatible with the data to be processed.
Contribution
Here we propose and demonstrate a method that does not use a set of training images or probabilistic atlases as priors. Instead we extract an ensemble of corresponding structures simultaneously. The evolving segmentation of the entire image set supports each of the individual segmentations. In practice, a subset of the model parameters, called the spatial parameters, is inferred as part of the joint segmentation processes. These latent spatial parameters, which can be viewed as a `dynamic atlas', are estimated exclusively from the data at hand and a single manual segmentation. The updated estimates of the latent atlas are used iteratively as Markov Random Field (MRF) priors on the tissue labels. The single node potentials term of the MRF model is formulated as a spatial constraint in a level-set functional for segmentation. The main novelty of the suggested method with respect to other group-wise segmentation methods is the consistent statistically-driven variational framework for MR ensemble segmentation by estimating a latent atlas.
Results
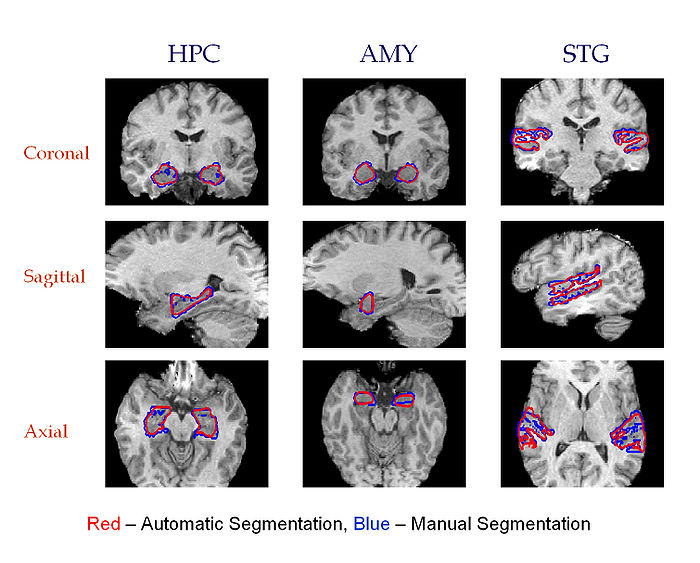
We test the proposed approach on 50 MR brain scans. Some of the subjects in this set are diagnosed with the first episode schizophrenia or affective disorder. The MR images (T1, 256X256X128 volume, 0.9375 X 0.9375 X 1.5 mm voxel size) were acquired by a 1.5-T General Electric Scanner. In addition to the MR volumes, manual segmentations of three structures (superior temporal gyrus, amygdala, and hippocampus) in each hemisphere were provided for each of the 50 individuals and used to evaluate the quality of the automatic segmentation results. MR images are preprocessed by skull stripping. The volumes were aligned using B-spline registration.
Three cross-sections of 3D segmentation of Hippocampus, Amygdala and Superior Temporal Gyrus in the left and the right hemispheres. Automatic segmentation is shown in red. Manual segmentation is shown in blue. Fourth column: Coronal views of the resulting atlases for each pair of structures.
Brain Tumor Modeling
We have applied the proposed algorithm to a longitudinal multi-modal Patient specific brain scans for brain tumor segmentation and modeling. In this particular application the inferred spatial parameters estimate the patient's latent anatomy. No prior information is assumed but a couple of mouse clicks that define a sphere that initializes the segmentation of the first time point. To learn more please refer to the brain tumor modeling page.
Publications
In Press
- T. Riklin Raviv, K. Van Leemput, W.M. Wells III and P. Golland, Joint Segmentation of Image Ensembles via Latent Atlases, MICCAI 2009.
- T. Riklin Raviv, B.H. Menze, K. Van Leemput, B. Stieltjes, M.A. Weber, N. Ayache, W. M. Wells III and P. Golland
Joint Segmentation using Patient specific Latent Anatomy Model, MICCAI workshop for Probabilistic Models on Medical Image Analysis (PMMIA) 2009
= Key Investigators
- MIT, CSAIL [| Tammy Riklin Raviv], Polina Golland
- Harvard Medical School, MGH | Koen Van Leemput
- Harvard Medical School, BWH | William M. Wells, Ron Kikinis, Martha Shenton, Sylvain Bouix