Difference between revisions of "DBP2:UNC:Local Cortical Thickness Pipeline"
From NAMIC Wiki
| Line 58: | Line 58: | ||
** Sulcal depth computation using genus zero surface and inflated one | ** Sulcal depth computation using genus zero surface and inflated one | ||
** Tool: MeshMath (UNC module) | ** Tool: MeshMath (UNC module) | ||
| − | * '''11. Cortical correspondence''' | + | * '''11. Particles initialization for cortical correspondence''' |
| + | ** Initializing particles on inflated surface using parcellation map and genus zero surface | ||
| + | ** Tools: ParticleInitializer (UNC Slicer3 external modules) | ||
| + | * '''12. Cortical correspondence''' | ||
** Correspondence on inflated surfaces using particle system | ** Correspondence on inflated surfaces using particle system | ||
** Tools: ParticleCorrespondencePreProcessing, ParticleCorrespondence, ParticleCorrespondencePostProcessing (UNC Slicer3 external modules) | ** Tools: ParticleCorrespondencePreProcessing, ParticleCorrespondence, ParticleCorrespondencePostProcessing (UNC Slicer3 external modules) | ||
| − | * ''' | + | * '''13. Group statistical analysis''' |
** Tool: QDEC Slicer module or StatNonParamPDM | ** Tool: QDEC Slicer module or StatNonParamPDM | ||
Revision as of 20:42, 7 December 2009
Home < DBP2:UNC:Local Cortical Thickness PipelineBack to UNC Cortical Thickness Roadmap
Contents
Objective
We would like to create end-to-end applications within Slicer3 allowing individual and group analysis of mesh-based local cortical thickness.
Pipeline overview
Input: RAW images (T1-weighted, T2-weighted, PD-weighted images)
- 1. Tissue segmentation
- Probabilistic atlas-based automatic tissue segmentation via an Expectation-Maximization scheme
- Tool: itkEMS (UNC Slicer3 external module)
- 2. Atlas-based ROI segmentation: subcortical structures, lateral ventricles, parcellation
- 2.1. Skull stripping using previously computed tissue segmentation label image
- Tool: SegPostProcess (UNC Slicer3 external module)
- 2.2. T1-weighted atlas deformable registration
- B-spline pipeline registration
- Tool: RegisterImages (Slicer3 module)
- 2.3. Applying transformations to the structures
- Tool: ResampleVolume2 (Slicer3 module)
- 2.1. Skull stripping using previously computed tissue segmentation label image
- 3. White matter map creation
- Brainstem and cerebellum extraction
- Adding subcortical structures except amygdala and hippocampus
- Tool: ImageMath (UNC Slicer3 external module)
- 4. White matter map post-processing
- Largest component computation
- Smoothing: Level set smoothing or weighted average filter
- Connectivity enforcement (6-connectivity)
- White matter filling
- Tool: WMSegPostProcess (UNC Slicer3 external module)
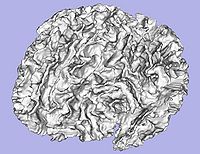
- 5. Genus zero white matter map image and surface creation
- Tool: GenusZeroImageFilter (UNC Slicer3 external module)
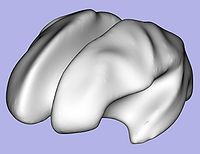
- 6. White matter surface inflation
- Iterative smoothing using relaxation operator (considering average vertex) and L2 norm of the mean curvature as a stopping criterion
- Iteration stopped if vertices that have too high curvature (some extremities)
- Tool: MeshInflation (UNC Slicer3 external module)
- 6 bis(Optional). White matter image fixing if necessary
- Correction of the white matter map image (corresponding to vertices that have high curvature) with connectivity enforcement
- Tool: FixImage (UNC Slicer3 external module)
- Go back to step 5
- 7. Gray matter map creation
- Adding genus zero white matter map to gray matter segmentation (without cerebellum and brainstem)
- Tool: ImageMath
- 8. Label map creation
- Label map creation for cortical thickness computation (WM + GM + CSF)
- Tool: ImageMath
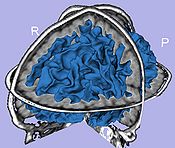
- 9. Cortical thickness
- Asymmetric local cortical thickness or Laplacian cortical thickness
- Tool: UNCCortThick or measureThicknessFilter (UNC Slicer3 external modules)
- 10. Sulcal depth
- Sulcal depth computation using genus zero surface and inflated one
- Tool: MeshMath (UNC module)
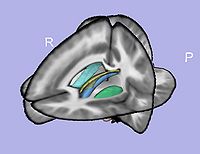
- 11. Particles initialization for cortical correspondence
- Initializing particles on inflated surface using parcellation map and genus zero surface
- Tools: ParticleInitializer (UNC Slicer3 external modules)
- 12. Cortical correspondence
- Correspondence on inflated surfaces using particle system
- Tools: ParticleCorrespondencePreProcessing, ParticleCorrespondence, ParticleCorrespondencePostProcessing (UNC Slicer3 external modules)
- 13. Group statistical analysis
- Tool: QDEC Slicer module or StatNonParamPDM
Download
Brain atlases
Three brain atlases are available on MIDAS and on NITRC:
- Pediatric atlas: http://www.insight-journal.org/midas/item/view/2277
- Adult atlas: http://www.insight-journal.org/midas/item/view/2328
- Elderly atlas: http://www.insight-journal.org/midas/item/view/2330
Pipeline validation
Analysis on a small pediatric dataset
Tests will be computed on a small pediatric dataset which includes 2 year-old and 4 year-old cases.
- 16 autistic cases
- 1 developmental delay
- 3 normal control
Comparison to state of the art
We would like to compare our pipeline with FreeSurfer. We will thus perform a regional statistical analysis using Pearson's correlation coefficient on an adult dataset (FreeSurfer's publicly available tutorial dataset) including 40 cases.
Planning
Done
Steps 1 to 10:
- Development of UNC Slicer3 modules (except itkEMS)
- Modules applied on small pediatric dataset from the Autism study
In progress
- Step 6: Parameter adjustment on autism dataset to fix bad vertices
- Step 11: Particle correspondence testing with pediatric surfaces
- Automatization of several steps
- Symmetric atlases generation (pediatric, adult, elderly):
- T1-weighted atlas
- Tissue segmentation probability maps
- Subcortical structures probability maps
Future work
- Full pipeline working on pediatric dataset
- Workflow for individual analysis as a Slicer3 high-level module using BatchMake
- Workflow for group analysis
References
- I. Oguz, M. Niethammer, J. Cates, R. Whitaker, T. Fletcher, C. Vachet, and M. Styner, Cortical Correspondence with Probabilistic Fiber Connectivity, Information Processing in Medical Imaging, IPMI 2009, LNCS, in print.
- H.C. Hazlett, C. Vachet, C. Mathieu, M. Styner, J. Piven, Use of the Slicer3 Toolkit to Produce Regional Cortical Thickness Measurement of Pediatric MRI Data, presented at the 8th Annual International Meeting for Autism Research (IMFAR) Chicago, IL 2009.
- C. Mathieu, C. Vachet, H.C. Hazlett, G. Geric, J. Piven, and M. Styner, ARCTIC – Automatic Regional Cortical ThICkness Tool, UNC Radiology Research Day 2009 abstract
- C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2009 abstract