Difference between revisions of "DBP2:UNC:Local Cortical Thickness Pipeline"
| Line 172: | Line 172: | ||
*I. Oguz, M. Niethammer, J. Cates, R. Whitaker, T. Fletcher, C. Vachet, and M. Styner, Cortical Correspondence with Probabilistic Fiber Connectivity, Information Processing in Medical Imaging, IPMI 2009, LNCS, in print. | *I. Oguz, M. Niethammer, J. Cates, R. Whitaker, T. Fletcher, C. Vachet, and M. Styner, Cortical Correspondence with Probabilistic Fiber Connectivity, Information Processing in Medical Imaging, IPMI 2009, LNCS, in print. | ||
*C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2010 abstract | *C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2010 abstract | ||
| − | |||
*C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2009 abstract | *C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2009 abstract | ||
*Oguz, I., Cates, J., Fletcher, T., Whitaker, R., Cool, D., Aylward, S., Styner, M., Cortical correspondence using entropy-based particle systems and local features, IEEE Symposium on Biomedical Imaging ISBI 2008. 1637– 1640 | *Oguz, I., Cates, J., Fletcher, T., Whitaker, R., Cool, D., Aylward, S., Styner, M., Cortical correspondence using entropy-based particle systems and local features, IEEE Symposium on Biomedical Imaging ISBI 2008. 1637– 1640 | ||
*J. Cates, P. Fletcher, M. Styner, H. Hazlett, R. Whitaker, Particle-based shape analysis of multi-object complexes, MICCAI 2008, 477-85 | *J. Cates, P. Fletcher, M. Styner, H. Hazlett, R. Whitaker, Particle-based shape analysis of multi-object complexes, MICCAI 2008, 477-85 | ||
*Cates, J., Fletcher, P., Whitaker, R.: Entropy-based particle systems for shape correspondence. MFCA Workshop, MICCAI 2006, 90–99 | *Cates, J., Fletcher, P., Whitaker, R.: Entropy-based particle systems for shape correspondence. MFCA Workshop, MICCAI 2006, 90–99 | ||
Revision as of 19:44, 26 August 2010
Home < DBP2:UNC:Local Cortical Thickness PipelineBack to UNC Cortical Thickness Roadmap
Contents
Objective
We would like to create an end-to-end application within Slicer3 allowing group-wise automatic mesh-based analysis of cortical thickness as well as other surface measurements (surface area...)
This page describes the related pipeline with its basic components, as well as its validation.
Pipeline overview
A Slicer3 high-level module for group-wise cortical thickness analysis has been developed: GAMBIT (Group-wise Automatic Mesh Based analysis of cortIcal Thickness)
Input: CSV file containing RAW images (T1-weighted, T2-weighted, PD-weighted images)
- 1. Individual pipeline
- 1.1. Tissue segmentation
- Probabilistic atlas-based automatic tissue segmentation via an Expectation-Maximization scheme
- Tool: itkEMS (UNC Slicer3 external module)
- 1.2. Atlas-based ROI segmentation: subcortical structures, lateral ventricles, parcellation
- 1.2.1. Skull stripping using previously computed tissue segmentation label image
- Tool: SegPostProcess (UNC Slicer3 external module)
- 1.2.2. T1-weighted atlas deformable registration
- B-spline pipeline registration
- Tool: RegisterImages (Slicer3 module)
- 1.2.3. Applying transformations to the structures
- Tool: ResampleVolume2 (Slicer3 module)
- 1.2.1. Skull stripping using previously computed tissue segmentation label image
- 1.3. White matter map creation
- Brainstem and cerebellum extraction
- Adding subcortical structures except amygdala and hippocampus
- Tool: ImageMath (UNC Slicer3 external module)
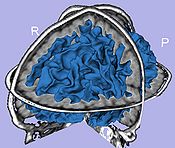
- 1.4. Cortical thickness computation
- Asymmetric cortical thickness
- Tool: UNCCortThick(UNC Slicer3 external module)
- 1.5. White matter map post-processing
- Largest component computation
- Smoothing: Level set smoothing or weighted average filter
- Connectivity enforcement (6-connectivity)
- White matter filling
- Tool: WMSegPostProcess (UNC Slicer3 external module)
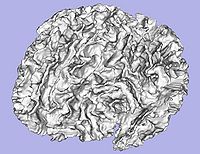
- 1.6. Genus zero white matter map image and surface creation
- Tool: GenusZeroImageFilter (UNC Slicer3 external module)
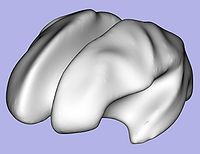
- 1.7. White matter surface inflation
- Iterative smoothing using relaxation operator (considering average vertex) and L2 norm of the mean curvature as a stopping criterion
- Iteration stopped if vertices that have too high curvature (some extremities)
- Tool: MeshInflation (UNC Slicer3 external module)
- 1.7 bis(Optional). White matter image fixing if necessary
- Correction of the white matter map image (corresponding to vertices that have high curvature) with connectivity enforcement
- Tool: FixImage (UNC Slicer3 external module)
- Go back to step 1.6
- 1.8. Sulcal depth
- Sulcal depth computation using genus-zero surface and inflated one
- Tool: MeshMath (UNC module)
- 1.9. Surface area computation
- Lobar surface area measurement on smoothed genus-zero surface
- Tool: MeshMath (UNC module)
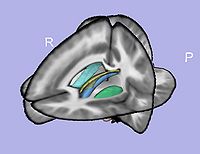
- 1.10. Particles initialization for cortical correspondence
- Initializing particles on inflated genus-zero surface using 98-lobe parcellation map and genus zero surface
- Tool: ParticleInitializer (UNC Slicer3 external module)
- 1.1. Tissue segmentation
- 2. Particle-based shape correspondence
- Correspondence on inflated surfaces using particle system
- 2.1. Preprocessing
- Distance maps creation from inflated genus-zero surfaces with slight gaussian blurring
- Tool: ParticleCorrespondencePreProcessing (UNC Slicer3 external module)
- 2.2. Correspondence optimization
- Particle-based shape correspondence optimization (using sulcal depth) with Procrustes alignement
- Tool: ShapeWorksRun (Utah Slicer3 external module)
- 2.3. Postprocessing
- Re-meshing using template
- Tool: ParticleCorrespondencePostProcessing (UNC Slicer3 external module)
- 2.4. Cortical thickness interpolation
- Cortical thickness interpolation on surface in correspondence
- Tool: MeshMath (UNC module)
- 3. Group statistical analysis
- Tool: QDEC Slicer module or StatNonParamPDM
GAMBIT Download
CVS access, Executables and tutorial dataset
Available on NITRC : http://www.nitrc.org/projects/gambit/
Brain atlases
Four brain atlases are available on MIDAS and on NITRC:
- Pediatric atlas: http://www.insight-journal.org/midas/item/view/2277
- Adult atlas: http://www.insight-journal.org/midas/item/view/2328
- Elderly atlas: http://www.insight-journal.org/midas/item/view/2330
- Primate atlas: http://www.insight-journal.org/midas/item/view/2283
Pediatric MRI Brain data
Data of 2 autistic children and 2 normal controls (male, female) scanned at 2 years with follow up at 4 years from a 1.5T Siemens scanner. Files include structural data, tissue segmentation label map and subcortical structures segmentation.
Pipeline validation
Analysis on a small pediatric dataset
Tests will be computed on a small pediatric dataset which includes 2 year-old and 4 year-old cases.
- 16 autistic cases
- 1 developmental delay
- 3 normal control
Comparison to state of the art
We would like to compare our pipeline with FreeSurfer. We will thus perform a regional statistical analysis using Pearson's correlation coefficient on an adult dataset (FreeSurfer's publicly available tutorial dataset) including 40 cases.
Planning
Done
- Workflow for group analysis (Slicer3 external module using BatchMake):
- Development of UNC Slicer3 modules
- Modules applied on small pediatric dataset from the Autism study
- Pediatric and adult brain atlases available to the community via MIDAS
- T1-weighted atlas
- Tissue segmentation probability maps
- Subcortical structures probability maps
- LBinary mask images
- GAMBIT available to the community via NITRC: executables (UNC external modules for Slicer3) and tutorial dataset
- Tutorial with application example on a small dataset
- GAMBIT source code (CVS) available to the community
In progress
- Step 1.7: Parameter exploration on autism dataset to improve inflation-fixing steps
- Step 2: Particle correspondence testing with pediatric surfaces (Meeting with Josh Cates at UNC - February 2010)
- New version of GAMBIT including quality control through MRML scene, and WM, GM models generation
- GAMBIT executable can be downloaded directly within Slicer3 using the extension wizard
References
- I. Oguz, M. Niethammer, J. Cates, R. Whitaker, T. Fletcher, C. Vachet, and M. Styner, Cortical Correspondence with Probabilistic Fiber Connectivity, Information Processing in Medical Imaging, IPMI 2009, LNCS, in print.
- C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2010 abstract
- C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2009 abstract
- Oguz, I., Cates, J., Fletcher, T., Whitaker, R., Cool, D., Aylward, S., Styner, M., Cortical correspondence using entropy-based particle systems and local features, IEEE Symposium on Biomedical Imaging ISBI 2008. 1637– 1640
- J. Cates, P. Fletcher, M. Styner, H. Hazlett, R. Whitaker, Particle-based shape analysis of multi-object complexes, MICCAI 2008, 477-85
- Cates, J., Fletcher, P., Whitaker, R.: Entropy-based particle systems for shape correspondence. MFCA Workshop, MICCAI 2006, 90–99