Difference between revisions of "TBISegmentation TutorialContestSummer2011"
| Line 5: | Line 5: | ||
by | by | ||
| − | '''Andrei Irimia''', '''Micah Chambers''' and '''Jack Van Horn''' | + | '''<span style="color:red">Andrei Irimia'''</span>, '''<span style="color:red">Micah Chambers'''</span> and '''<span style="color:red">Jack Van Horn'''</span> |
UCLA Laboratory of Neuro Imaging (http://loni.ucla.edu/). | UCLA Laboratory of Neuro Imaging (http://loni.ucla.edu/). | ||
Revision as of 20:36, 3 June 2011
Home < TBISegmentation TutorialContestSummer2011AUTOMATIC SEGMENTATION OF TRAUMATIC BRAIN INJURY MRI VOLUMES USING ATLAS BASED CLASSIFICATION AND 3D SLICER
by
Andrei Irimia, Micah Chambers and Jack Van Horn
UCLA Laboratory of Neuro Imaging (http://loni.ucla.edu/).
Contents
Background
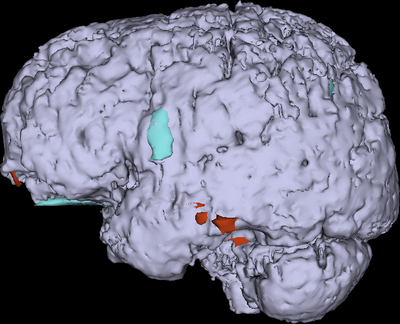
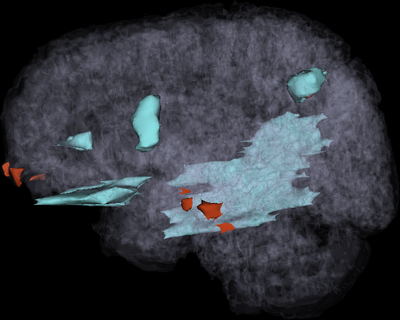
Traumatic brain injury (TBI) is associated with an estimated 1.7 million cases in the United States alone every year, with an estimated 1.2 million ER visits and over 50,000 deaths. Over 5.3 million cases of required long‐term daily assistance exist as a result of TBI, which results in health care costs of over $60 billion/year. Interest in and public awareness of TBI-caused diffuse axonal injury (DAI) has surged with the publication of studies suggesting that DAI in professional and amateur athletes may have both acute and chronic effects upon neurocognitive function. Moreover, the large number of recent TBI cases in soldiers returning from military conflicts has created a significant clinical challenge for US Veterans Administration hospitals and has highlighted the critical need for improvement in TBI care and treatment. Prompt and proper management of TBI sequelae can significantly alter their course, improve mortality and morbidity, reduce hospital stay and decrease health care costs. Neuroimaging of TBI is thus vital for surgical planning by providing important information for anatomic localization and surgical navigation, as well as for guiding decisions regarding the aggressiveness of TBI treatment. Moreover, because TBI often results in characteristic impairments dependent on the area of involvement, lesion analysis can assist in the localization of cognitive processes and in the attempt to obtain novel information on the relationship between brain anatomy and behavior. For all these reasons, reliable and precise methods of TBI assessment can play an essential role during both acute and chronic therapy of this condition. Despite many innovations, progress towards patient‐tailored characterization of the structural and functional substrates associated with TBI‐related neural and cognitive impairment remains dissatisfactory and the relationship between neurophysiological markers of cognitive dysfunction and TBI structural damage has not been acceptably elucidated. 3D Slicer offers a powerful and unparalleled set of tools for the exploration and quantification of TBI.
Purpose
This tutorial has four purposes, namely it allows the user to
- acquire familiarity with several MR sequences commonly used for TBI imaging
- learn how to apply ABC to perform joint co-registration and automatic segmentation of TBI volumes
- acquire expertise on how to identify and characterize TBI pathology using various MRI sequences
- gain exposure to informed strategies for quantification of TBI-related edema or hemorrhage
Value of the tutorial
This tutorial demonstrates that 3D Slicer offers powerful methodologies for the visualization of pathology due to traumatic brain injury. Use of multiple MR image channels greatly enhances the ability to study and understand TBI structure/extent, and ABC is shown to be a robust algorithm to perform joint co-registration and automatic segmentation of TBI. Furthermore, completion of this tutorial allows one to acquire useful expertise on how to identify and characterize TBI. Use of 3D Slicer can offer informed strategies for quantification of TBI-related edema or hemorrhage and for improved insight of clinical relevance.
Prerequisites
Please note that the tutorial assumes that you are using Slicer 3.6 or later, and that you have already completed the Visualization and the Interactive Editor Tutorials by Sonia Pujol. The tutorial has been developed using Windows 7, but it is also compatible with Windows XP, Linux 32 and 64 bit, as well as Mac Darwin.
Tutorial and Data
Download times may vary due to the presence of three-dimensional animations of TBI in the tutorial.
Team Members and Mentors
- Andrei Irimia, Ph.D. (Postdoctoral Scholar, Laboratory of Neuro Imaging, UCLA)
- Micah C Chambers, MS (Graduate Student in Bioengineering, UCLA)
- John D Van Horn, M.Eng., Ph.D. (Assistant Professor of Neurology, UCLA)
- Paul M. Vespa, M.D., F.A.A.N., F.A.C.N. (Professor of Neurology and Neurosurgery, Director of the Brain Injury Research Center, UCLA)
- Arthur W. Toga, Ph.D. (Distinguished Professor of Neurology, Director of LONI, Co-Director of Brain Mapping Division, Associate Dean of the Medical School, UCLA)
- Jeffry R. Alger, Ph.D. (Professor of Radiological Sciences in Residence, UCLA)
- David A. Hovda, Ph.D. (Professor of Neurosurgery, UCLA)