Difference between revisions of "DBP3:Utah:AutoEndoSeg"
From NAMIC Wiki
| Line 33: | Line 33: | ||
|} | |} | ||
| + | = Larger Atlas Images = | ||
| + | * A second atlas was generated from cropped images that included a larger region of the initial MRI scans (namely, the appendage and a greater extent of the PVs was included in this second atlas). | ||
| + | * This atlas showed no marked improvement over the initial atlas | ||
| + | * Segmentations with the second atlas tended to not capture the entire blood pool, whereas, the first atlas tended to overestimate the blood pool. | ||
| + | * Registration issues? | ||
| − | = | + | {| class="wikitable" |
| + | |- | ||
| + | ! | ||
| + | ! Mask and LGE Image | ||
| + | ! Mask and Expert-defined Segmentation | ||
| + | |- | ||
| + | | Example 1 | ||
| + | | [[File:CARMA_N24_LGE.png|thumb|center]] | ||
| + | | [[File:CARMA_N24_Endo.png|thumb|center]] | ||
| + | |- | ||
| + | | Example 2 | ||
| + | | [[File:CARMA_N25_LGE.png|thumb|center]] | ||
| + | | [[File:CARMA_N25_Endo.png|thumb|center]] | ||
| + | |- | ||
| + | | Example 3 | ||
| + | | [[File:CAMRA_N26_LGE.png|thumb|center]] | ||
| + | | [[File:CARMA_N26_Endo.png|thumb|center]] | ||
| + | |- | ||
| + | | Example 4 | ||
| + | | [[File:CAMRA_N26_2_LGE.png|thumb|center]] | ||
| + | | [[File:CARMA_N26_2_Endo.png|thumb|center]] | ||
| + | |- | ||
| + | | | ||
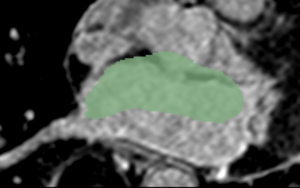
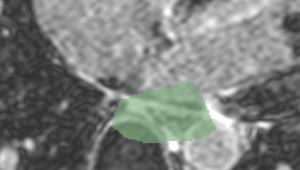
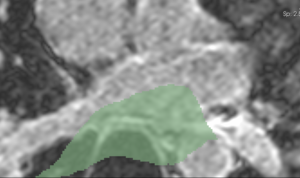
| + | | Cropped MRI image overlaid with the algorithm-defined blood pool. | ||
| + | | Expert manual segmentations (white) overlaid with the algorithm-defined blood pool (green). | ||
| + | |- | ||
| + | |} | ||
Revision as of 08:49, 29 November 2011
Home < DBP3:Utah:AutoEndoSegContents
Notes on the Endocardial Segmentation Algorithm
Algorithm is a multi-atlas segmentation.
- Register (cropped) heart region of patient DEMRI to all of the n atlas DEMRI images and record n minimum metric values (mutual information)
- Transform all n atlas segmentations to patient DEMRI space
- Segmentation is a weighted average of the transformed atlas segmentation, where weights are determined by the mutual information values from step 1
- Pick a threshold as your segmentation
Notes on Yi's Slicer Module
- The exposed parameter is the ratio of pixel samples for mutual information to the total number of pixels in the image.
- We could also expose the threshold parameter in the final step
- We can change or add atlas datasets by copying into the appropriate directory and editing the text file that lists them
- Uses the ITK mutual information code. Does affine registration followed by b-spline
- We will partner with Yi for module development at the Jan 2012 project week
Initial Experiences
- Comparison of GA Tech and Utah-derived segmentations
(click to enlarge)
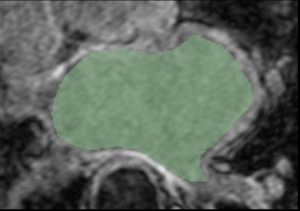
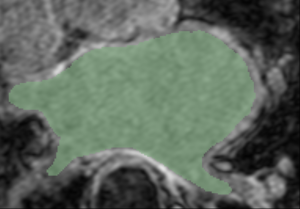
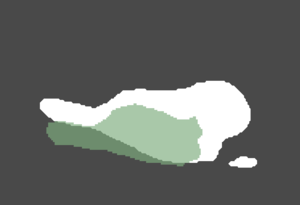
| GA Tech Segmentation | Utah Segmentation | Thresholding Difference |
|---|---|---|
| Segmentation of an atlas image by GA Tech, using their endocardial auto-segmentation algorithm. | Segmentation of the same image with the same algorithm by Utah. | User-based variability in thresholding the probability mask output. (Green region - Utah; white region - GA Tech). |
Larger Atlas Images
- A second atlas was generated from cropped images that included a larger region of the initial MRI scans (namely, the appendage and a greater extent of the PVs was included in this second atlas).
- This atlas showed no marked improvement over the initial atlas
- Segmentations with the second atlas tended to not capture the entire blood pool, whereas, the first atlas tended to overestimate the blood pool.
- Registration issues?
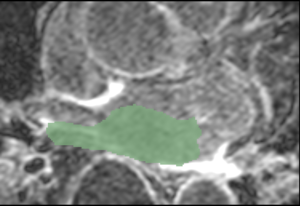
| Mask and LGE Image | Mask and Expert-defined Segmentation | |
|---|---|---|
| Example 1 | ||
| Example 2 | ||
| Example 3 | ||
| Example 4 | ||
| Cropped MRI image overlaid with the algorithm-defined blood pool. | Expert manual segmentations (white) overlaid with the algorithm-defined blood pool (green). |