Difference between revisions of "Projects:RegistrationLibrary:RegLib C33 v36"
From NAMIC Wiki
| Line 13: | Line 13: | ||
**[[Media:RegLib_C33_Presets.mrml|'''RegLib_C33_Presets''' Presets for FLAIR and DTI registration & DTI resampling <small> (.mrml file 12 kB) </small>]]''' | **[[Media:RegLib_C33_Presets.mrml|'''RegLib_C33_Presets''' Presets for FLAIR and DTI registration & DTI resampling <small> (.mrml file 12 kB) </small>]]''' | ||
**[[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]] | **[[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]] | ||
| + | === Overall Strategy === | ||
| + | :#DWI -> DTI, obtain mask +baseline | ||
| + | :#Subsample T1 to isotropic 1x1x1 mm -> T1_sub | ||
| + | :#Register DTI_baseline to T1_sub (affine+nonrigid) w/o masking | ||
| + | :#Resample DTI_mask with above transform to obtain mask for T1_sub | ||
| + | :#Re-register DTI_baseline to T1_sub (affine+nonrigid) '''with''' masking | ||
| + | :#Resample DTI with result nonrigid transform | ||
| + | :#Resample DTI_mask with above nonrigid transform and FLAIR as reference -> yields mask for FLAIR | ||
| + | :#Resample DTI_mask with above nonrigid transform and T1Gd as reference -> yields mask for T1Gd | ||
| + | :#Register FLAIR and T1Gd to T1_sub (affine only) '''with''' masking | ||
=== Procedures === | === Procedures === | ||
**'''Phase I: Load & Convert DICOM''' | **'''Phase I: Load & Convert DICOM''' | ||
Revision as of 19:22, 16 May 2012
Home < Projects:RegistrationLibrary:RegLib C33 v36Contents
RegLib C36 Procedures for Slicer 3.6
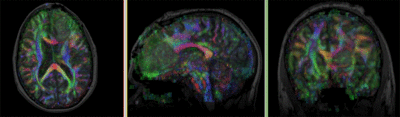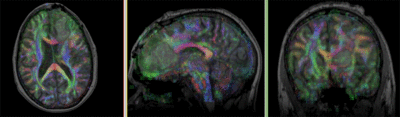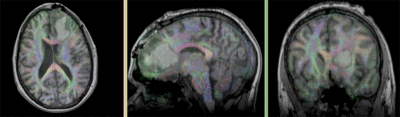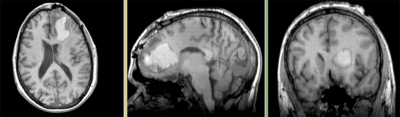
Registration Results
Download
- Data
- RegLib_C33_Data DWI, DTI, T1, FLAIR, Presets & solution transforms (NRRD files, zip file 174 MB)
- RegLib_C33_Data_noDWI subset of the above, contains DTI but w/o DWI and resampled DTI (NRRD files, zip file 86 MB)
- RegLib_C33_Data Transforms & Presets only contains only result transforms and parameter presets (NRRD files, zip file 30 kB)
- Presets
Overall Strategy
- DWI -> DTI, obtain mask +baseline
- Subsample T1 to isotropic 1x1x1 mm -> T1_sub
- Register DTI_baseline to T1_sub (affine+nonrigid) w/o masking
- Resample DTI_mask with above transform to obtain mask for T1_sub
- Re-register DTI_baseline to T1_sub (affine+nonrigid) with masking
- Resample DTI with result nonrigid transform
- Resample DTI_mask with above nonrigid transform and FLAIR as reference -> yields mask for FLAIR
- Resample DTI_mask with above nonrigid transform and T1Gd as reference -> yields mask for T1Gd
- Register FLAIR and T1Gd to T1_sub (affine only) with masking
Procedures
- Phase I: Load & Convert DICOM
- run Dicom2NRRD converter for the DWI Dicom set. Output file: DWI.nrrd
- load DWI.nrrd
- load T1, FLAIR and T1Gd images via AddVolume...
- Subsample T1: The T1 is 512 x 512 x 176 (0.5 x 0.5 x 1) resolution, which is too large as a reference to sample the DTI into. So we generate a smaller, isotropic 1x1x1 version of the T1 that can serve as reference:
- Open Filtering/Resample Scalar Volume module
- Input: T1, Output: new volume, rename to T1_sub
- interpolation: Hamming
- Spacing: 1,1,1
- Phase II: Check initial Alignment and anisotropy
- large initial misalignment and high voxel anisotropy can cause problems when registering and resampling the DTI. In such cases it is advisable to resample the DWI first into a more isotropic space and, if necessary, initially realign to a similar orientation as the T1 reference, 'before converting to DTI. In this example DWI is sufficiently aligned and has voxel dimensions of 1 x 1 x 2.6, which is borderline. Anisotropy ratios greater than 1:3 tend to benefit from initial resampling. In this case we proceed without.
- 'Phase III: convert DWI -> DTI
- go to module: Diffusion / Utilities / Diffusion Tensor Estimation
- input: DWI, output : DTI, DTI_base and DTI_mask
- Input DWI Volume: DWI, Output DTI Volume: create new, rename to "DTI"
- Output Baseline: create new, rename to "DTI_base"
- Otsu Threshold Mask: create new, rename to "DTI_iso_mask"
- check boxes for Remove Islands and Apply Mask
- leave defaults
- Phase IVa: Register DTI-T1 (unmasked)
- open Registration : BrainsFit module (presets: Xf1_DTI-T1_unmasked
- Registration Phases:
- set T1_sub as fixed and DTI_base as moving image
- select/check Include Affine registration phase
- select/check Include BSpline registration phase
- Output Settings:
- select a new transform "Slicer BSpline Transform", rename to "Xf1_DTI-T1_unmasked"
- select a new volume "Output Image Volume, rename to "DTI_base_Xf1"
- Registration Parameters: increase Number Of Samples to 200,000
- Registration Parameters: set Number Of Grid Subdivisions to 5,5,3
- Leave all other settings at default
- click: Apply; (runtime < 1 min 10 sec. on MacPro Quad Core 2.4Ghz)
- result: the absence of the skull in the DTI causes significant and incorrect stretch of image in the I-S direction.
- we repeat with mask, using the above estimate to produce a mask for the T1
- Phase IVb: Build mask for T1: resample DTI_mask
- go to Filtering/Resample Scalar Vector DWI Volume
- Use provided presets or set the following:
- input: DTI_mask
- reference: T1_sub
- output: create new, rename to T1_sub_mask
- Transform node: Xf1_DTI-T1_unmasked
- Transforms order: check output-to-input
- interpolation type: check "nn"
- click: Apply
- Go to Volumes module
- select new "T1_sub_mask: volume, go to Info tab
- check the labelmap checkbox
- Phase IVc: Register DTI-T1 (masked)
- open Registration : BrainsFit module (presets: Xf1_DTI-T1_masked)
- settings as in Phase IVa above, except:
- select a new transform "Slicer BSpline Transform", rename to "Xf2_DTI-T1_masked"
- select a new volume "Output Image Volume, rename to "DTI_base_Xf2"
- Mask Processing Mode: check "ROI"
- Input Fixed Mask: T1_mask
- Input Moving Mask: DTI_mask
- Apply. Runtime 45 sec.
- go to Volumes module to adjust window&level for new result "DTI_base_Xf2"
- alignment is now much improved
- settings as in Phase IVa above, except:
- Phase V: Resample DTI
- Open the Resample DTI Volume module (found under: All Modules)
- Input Volume: select DTI
- Output Volume: select New DTI Volume, rename to DTI_Xf2
- Reference Volume: select T1_sub
- Transform Parameters: select transform "Xf2_DTI-T1_masked
- check box: output-to-input
- Leave all other settings at defaults
- Click Apply; runtime ~ 3 min.
- Phase VIa: Register FLAIR & T1 Gd: obtain masks
- we seek affine alignment only for FLAIR and T1Gd. As above, better results are achieved when masking non-brain parenchyma. To obtain masks for the FLAIR and T1Gd we proceed as above by resampling the DT_mask with the Xf2_DTI-T1 transform and the respective image as reference.
- go to Filtering/Resample Scalar Vector DWI Volume
- input: DTI_mask
- reference: FLAIR
- output: create new, rename to FLAIR_mask
- Transform node: Xf2_DTI-T1_masked
- Transforms order: check output-to-input
- interpolation type: check "nn"
- click: Apply
- Go to Volumes module
- select new "FLAIR_mask: volume, go to Info tab
- check the labelmap checkbox
- Phase VIb: Mask for T1Gd
- ditto: we seek affine alignment only for T1Gd. Identical procedure as for FLAIR above:
- go to Filtering/Resample Scalar Vector DWI Volume
- input: DTI_mask
- reference: T1Gd
- output: create new, rename to T1Gd_mask
- Transform node: Xf2_DTI-T1_masked
- Transforms order: check output-to-input
- interpolation type: check "nn"
- click: Apply
- Go to Volumes module
- select new "T1Gd_mask: volume, go to Info tab
- check the labelmap checkbox
- Phase VIc: Register FLAIR & T1Gd
- open Registration : BrainsFit module (presets: Xf4_... and Xf5_...)
- Registration Phase: check box for "Affine" only
- select a new linear transform "Slicer Linear Transform", rename to "Xf4_FLAIR-T1_masked"
- Mask Processing Mode: check "ROI"
- Input Fixed Mask: T1_mask
- Input Moving Mask: FLAIR_mask
- Apply. Runtime ~45 sec.
- Registration Phase: check box for "Affine" only
- ditto for T1Gd