Difference between revisions of "2012 Summer Project Week:Bladder Segmentation in Slicer"
From NAMIC Wiki
| Line 35: | Line 35: | ||
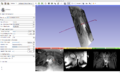
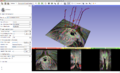
Tools previously mentioned were successfully used to segment out the bladder from MRI images, and 3D images were rendered as a result. Three tutorial modules were created outlining the mechanism of execution, included in this page. Further segmentation efforts will be conducted on different data sets. | Tools previously mentioned were successfully used to segment out the bladder from MRI images, and 3D images were rendered as a result. Three tutorial modules were created outlining the mechanism of execution, included in this page. Further segmentation efforts will be conducted on different data sets. | ||
| − | [[Media:Combined_Cropping_Growcut_and_Draw_Tool_Tutorial_for_Bladder_Imaging. | + | [[Media:Combined_Cropping_Growcut_and_Draw_Tool_Tutorial_for_Bladder_Imaging.doc|Combined GrowCut, Crop, and Draw Turotial]] |
</div> | </div> | ||
Revision as of 16:19, 25 June 2012
Home < 2012 Summer Project Week:Bladder Segmentation in SlicerKey Investigators
- UMass Amherst: Scott Tyler Blevins
- BWH: Scott Tyler Blevins, Nabgha Farhat, Jan Egger, Tobias Penzkofer, Tina Kapur
Objective
The goal of this project is to investigate efficient (less than 30s) methods for segmentation of bladder from MRI images of the female pelvis in Slicer.
Approach, Plan
We will explore different segmentation/editing tools in Slicer to determine which ones can most efficiently segment the bladder from MRI images of the female pelvis. Tools anticipated use include Growcut, Draw, and Crop.
Progress
Tools previously mentioned were successfully used to segment out the bladder from MRI images, and 3D images were rendered as a result. Three tutorial modules were created outlining the mechanism of execution, included in this page. Further segmentation efforts will be conducted on different data sets.
Delivery Mechanism
N/A