Whole-Brain-Tractography-Wizard
The Idea of a Wizard
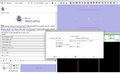
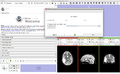
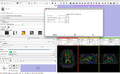
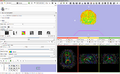
Make a simple set of steps that will guide the user through a complex process. An example of this is the simple wizard that takes the user from a DICOM or NRRD Diffusion Weighted Image to a Full brain tractography. There are 4 steps to this process:
In order to generate this wizard, we will use the For each step, we must:
- Obtain the input from the interface
- Validate the input
- Set up the parameters for the next step
API support for workflows
In order to do this, we use CTK's support for workflows:
- ctkWorkflowStackedWidget
- ctkWorkflowWidgetStep
In which the idea is to derive a new class from ctkWorkflowWidgetStep for each step and then add them all to the workflow widget ctkWorkflowStackedWidget making the transition explicit
Events of each workflow widget step (ctkWorkflowWidgetStep)
- createUserInterface
- onEntry(comingFrom, transitionType)
- onExit(goingTo, transitionType)
- validate(validationSucceded, desiredBranchId)
Obtaining the input from the Interface
You can find the example at
Modules/Scripted/Scripts/DICOM2FullBrainTractography/DICOM2FullBrainTractographyLib/full_tractography_workflow.py
Declaring the Modules
step_widget_files = [
'dicom2nrrd',
'dwi2dti',
'dti2fibers',
'done',
]
Declaring Each Module's Fields
step_widget_fields = {
'dicom2nrrd':[
('DICOMRadioButton', 'checked'),
('NRRDDWIRadioButton', 'checked'),
('inputDicomDirectory', 'directory'),
('outputVolume', 'currentPath'),
('useBMatrixGradientDirections','checked'),
('inputNRRDVolume','currentPath'),
],
'dwi2dti':[
('leastSquaresEstimation', 'checked'),
('weightedLeastSquaresEstimation', 'checked'),
('thresholdParameter', 'value'),
('removeIslands', 'checked'),
('applyMask', 'checked'),
],
'dti2fibers':[
('seedSpacing','value'),
('stoppingFAValue','value'),
('minimumFAValueSeed','value'),
('stoppingTrackCurvature','value'),
],
'done':[],
}
Validating Data for a Field
def validate_dicom2nrrd(self, step_object, data):
if data[step_object.id()]['DICOMRadioButton']:
Running a CLI module from a python script
self.dicomtonrrdconverter_parameter_node = slicer.cli.run(
slicer.modules.dicomtonrrdconverter, self.dicomtonrrdconverter_parameter_node,
data[step_object.id()],
wait_for_completion = True)
Validating the result of a CLI module
if self.dicomtonrrdconverter_parameter_node.GetStatusString() == 'Completed':
file_path = data[step_object.id()]['outputVolume']
result_status, node = slicer.util.loadVolume(
file_path,
True
)
else:
result_status = False
Setting data for the next module
if result_status:
self.dwi_node = node
self.dwi_node_name = node.GetID()
Output errors if needed
if not result_status:
display_error("Error in DICOM to NRRD conversion, please see log")
return result_status
Details on Each Step's CallBack Implementation
Entry Callback
def onEntry(self, comingFrom, transitionType):
comingFromId = "None"
if comingFrom: comingFromId = comingFrom.id()
super(GeneralizedStep, self).onEntry(comingFrom, transitionType)
if hasattr(self, 'onEntryCallback'):
self.onEntryCallback(self, comingFrom, transitionType)
Exit Callback
def onExit(self, goingTo, transitionType):
goingToId = "None"
if goingTo: goingToId = goingTo.id()
super(GeneralizedStep, self).onExit(goingTo, transitionType)
if hasattr(self, 'onExitCallback'):
self.onExitCallback(self, goingTo, transitionType)
Validation Callabck
def validate(self, desiredBranchId):
validationSuceeded = True
if hasattr(self, 'validateCallback'):
validationSuceeded = self.validateCallback(self, desiredBranchId)
super(GeneralizedStep, self).validate(validationSuceeded, desiredBranchId)