2014 Project Week:LongitudinalDTI
Key Investigators
- Utah: Anuja Sharma, Bo Wang, Guido Gerig
- UCLA: Andrei Irimia, John D. Van Horn
- UNC: Martin Styner
Project Description
Objective
- Patient specific modeling of longitudinal changes in white matter integrity along tracts, with emphasis on seemingly unaffected regions of the brain.
- Identify changes in fiber tract architecture along with possible WM atrophy reflected in the evolution of diffusion properties.
- Specific goals for this week include DTI pre-processing to improve the quality of the tractography results that we have obtained in the past for our dataset.
Approach, Plan
- Quality check of the TBI data using DTIPrep to correct for DTI related imaging artifacts, including checking for potential bias fields.
- Validation of the estimated DTI via glyph visualizations.
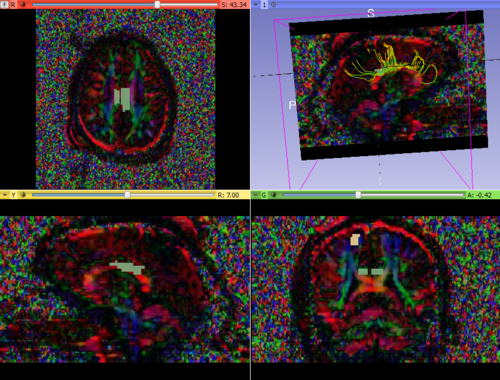
- Longitudinal co-registration of the patient's scans followed by tractography to verify the consistency of the extracted white matter tracts.
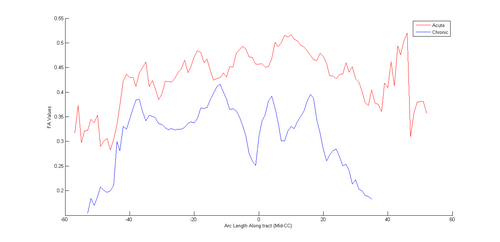
- Comparison of diffusion profiles along tract in a longitudinal setup to identify possible changes at the chronic timepoint.
Progress
- The proposed workflow was successfully carried out in Slicer for a single TBI patient with two longitudinal DTI scans. The detailed steps are described in the following sections.
- Utilized components of the UNC-Utah NAMIC tools for the current analysis.
- Had discussions with the UCLA TBI team on future directions, attended the TBI DBP meeting and the DTI breakout session.
Patient-specific longitudinal DTI analysis
The preliminary focus was to improve the data quality via DTI pre-processing. We tried out various options including rician noise filtering for DWI smoothing, checking for possible bias fields, common DTI related artifacts like motion correction and eddy current artifacts. Overall, we got the best results by going through the complete DTIPrep pipeline for both the longitudinal scans. The acute scan saw considerable quality improvement after the DTI Prep QC. For the sanity check of the tensors estimated from the corrected images, we used the glyph visualizations. However since a TBI patient may exhibit several types of lesions and edema with varying levels of severity, the actual white matter pathways may be damaged and/or bent out of shape. Since the degree of white matter integrity being compromised is usually unknown, we found the glyphs alone were hard to validate visually at both timepoints.