Projects:fMRIClustering
Back to NA-MIC_Collaborations, MIT Algorithms
fMRI Clustering
In this work we propose to simultaneously estimate the optimal representative time courses that summarize the fMRI data well and the partition of the volume into a set of disjoint regions that are best explained by these representative time courses. Our approach offers two advantages. First, is removes the sensitivity of the analysis to the details of the seed selection. Second, it substantially simplifies group analysis by eliminating the need for a subject-specific threshold at which correlation values are deemed significant. This unsupervised technique generalizes connectivity analysis to situations where candidate seeds are di�cult to identify reliably or are unknown. Our experimental results indicate that the functional segmentation provides a robust, anatomically meaningful and consistent model for functional connectivity in fMRI.
Description
Fiber Tractography
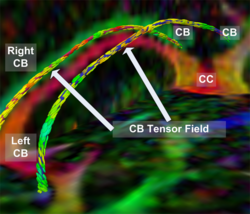
The goal of fiber tractography is to find open curves (i.e. paths without volume) in the white matter which correspond to something anatomically significant. In our framework, a direction-dependent Finsler manifold is constructed. Note that Riemannian manifolds based on tensors comprise a subset of all possible Finsler manifolds and are most commonly used in this framework. Then, the best geodesic path connecting the input ROIs is computed on the manifold. This minimum cost geodesic curve is determined by solving a Hamilton-Jacobi-Bellman (HJB) equation using the Fast Sweeping algorithm. The resulting solution is the anchor tract connecting the two ROIs.
Clinically speaking, the anchor tract has little meaning. However, during the computation of the anchor tract, connectivity maps are generated as a useful by-product which represent the connectivity of the brain volume to the input ROIs.
Also, note that in this framework, special care is taken so that the minimum cost (or equivalently the fastest travel time of a particle moving along the curve in the given manifold) and the anisotropic front propagation used to solve the HJB equation are appropriately related through a Legendre transformation. Algorithms which inappropriately ignore the Legendre transformation can severely impact the anisotropy information in front propagation based PDE solvers.
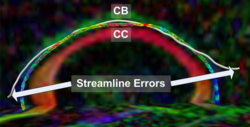
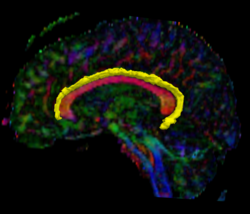
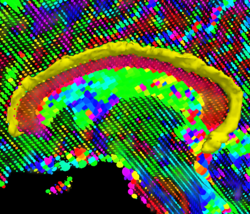
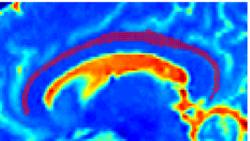
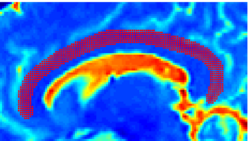
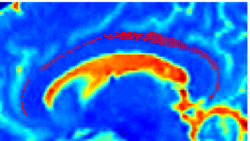
Recently, we have applied this method to the cingulum bundle, as shown in the following images:
Fiber Bundle Segmentation
We have developed a region-based active contour segmentation algorithm which provides volumetric fiber segmentations of fiber bundles. These active contours are initialized on the anchor tract and are expanded away from the anchor tract to capture the volumetric fiber bundle. The basic idea is to evolve the front away from the contour capturing only those voxels which are locally "connected" to the anchor tract. Cast in a Bayesian framework, this technique includes prior clinical knowledge penalizing bundles with abnormal radii and image-driven likelihood information to accurately capture edges in the directional information.
Data
We are using Harvard's high angular resolution datasets which currently consist of a population of 12 schizophrenics and 12 normal controls.
Associated Results
For previous results showing comparisons of this framework to traditional streamline frameworks and results showing the application of this framework to the segmentation of blood vessels, see Associated Results.
Statistical Results
We are currently investigating Cingulum Bundle fractional anisotropy (FA) differences between a population of 12 schizophrenics and 12 normal controls. We find the anchor tracts as described above and then compute statistics for FA inside a tube of radii 1-3mm centered on the anchor tract. So far using this method we have been unable to find a statistical difference between the normal controls and the schizophrenics. Once we have processed the entire population using our volumetric segmenter, we will rerun the statistics on the full fiber bundle.
Download the current statistical results here. (last updated 18/Apr/2007)
Project Status
- Working 3D implementation in Matlab using the C-based Mex functions.
- Currently porting to ITK.
Key Investigators
- Georgia Tech: John Melonakos, Vandana Mohan, Sam Dambreville, Allen Tannenbaum
- Harvard/BWH: Marek Kubicki, Marc Niethammer, Kate Smith, C-F Westin, Martha Shenton
Publications
In print
In press
- J. Melonakos, M. Niethammer, V. Mohan, M. Kubicki, J. Miller, and A. Tannenbaum. Locally-Constrained Region-Based Methods for DW-MRI Segmentation. MMBIA 2007.
- V. Mohan, J. Melonakos, M. Niethammer, M. Kubicki, and A. Tannenbaum. Finsler Level Set Segmentation for Imagery in Oriented Domains. BMVC 2007.
- J. Melonakos, V. Mohan, M. Niethammer, K. Smith, M. Kubicki, and A. Tannenbaum. Finsler Tractography for White Matter Connectivity Analysis of the Cingulum Bundle. MICCAI 2007 (accepted for oral presentation).
- J. Melonakos, E. Pichon, S. Angenet, and A. Tannenbaum. Finsler Active Contours. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2007.
Links
- For some additional diffusion-related thoughts, see: DW-MRI Musings
Project Week Results: Jan 2007, June 2007