2015 Summer Project Week:T1 mapping
From NAMIC Wiki
Home < 2015 Summer Project Week:T1 mapping
Key Investigators
- Xiao Da (MGH), Yangming Ou (MGH), Andriy Fedorov (BWH), Jayashree Kalpathy-Cramer (MGH)
- Utsav Pardasani (Observing)
Project Description
T1 mapping estimates effective tissue parameter maps (T1) from multi-spectral FLASH MRI scans with different flip angles. T1 mapping can be used to optimize parameters for a sequence, monitor diseased tissue, measure Ktrans in DCE-MRI and etc.
Objective
- Estimate effective T1 from multi-spectral FLASH MRI scans with different flip angles
- Implement T1 mapping algorithm as a Slicer module using C++
Approach, Plan
- Start with prostate diffusion module
- Update equations for T1 mapping
- Compare the results using C++, Python, Freesurfer with the ground truth of T1 for QIBA phantom
- Test the Slicer module on MGH Brain Tumor MR Data with multiple flip angles
Progress
- Updated the equation for variable flip angle (VFA) T1 mapping
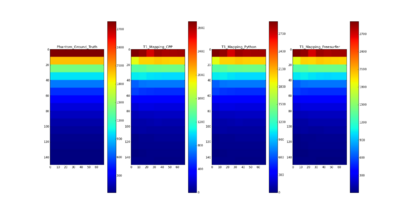
- Compared the results using C++ with Python, Freesurfer and the ground truth of T1 for QIBA phantom data
- T1 mapping results on QIBA phantom data using C++ are comparable with the ground truth and the results using Freesurfer and Python
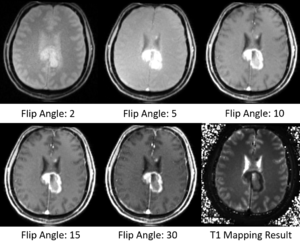
- Did some tests on MGH Brain Tumor MR Data with multiple flip angles
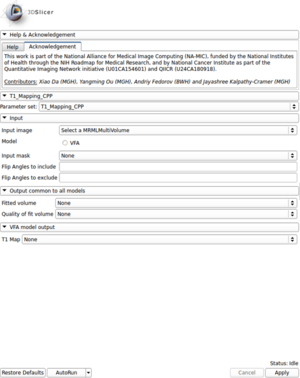
- Created a module for T1 mapping using C++
- Take multi-spectral FLASH images with an arbitrary number of flip angles as input, and estimate the T1 values of the data for each voxel
- Read repetition time(TR), echo time(TE) and flip angles from the Dicom header automatically
- Allow user to choose which flip angles will be included or excluded for the fitting process
- Output the fitting volume and quality of fitting image as well
- Uploaded the source code on Github