Slicer3:VisualBlog
- June through October 2007
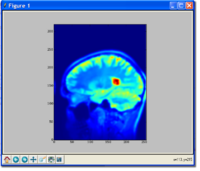
Image from Dirk, Matthew, Jim, and Steve on Monday, August 13, 2007
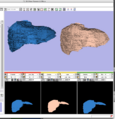
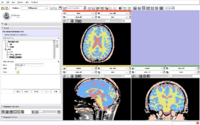
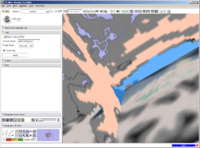
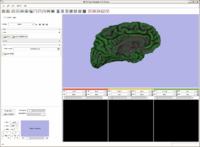
A new label map smoothing tool has been added to help with our collaboration with Children's Hospital Boston, SCI at University of Utah and Northeastern University. The unfiltered labelmap is shown in blue, and the filtered results are shown in peach. The filtering algorithm was developed at University of Utah by Josh Cates and Ross Whitaker, and implemented in ITK by GE Research. It is integrated in slicer3 as a command line module using the execution model. The models are shown unsmoothed in order to better visualize the underlying labelmaps.Image from Brad and Kilian on Wednesday, June, 21, 2007
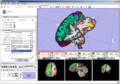
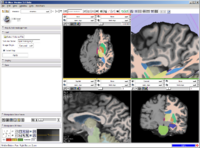
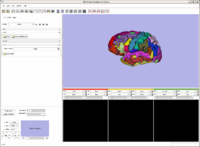
Example of EMSegmenter in Slicer3
- through June 2007
Image from ipek on Wednesday, June, 6, 2007
UNC Logo in Slicer3 for KWMeshVisu (Ipek Oguz, Martin Styner).
Image from wjp on Wednesday, May, 30, 2007
Design mockups for Slicer3's Cine Display interface (William Leue, Wendy Plesniak).
Image from pieper on Tuesday, May, 30, 2007
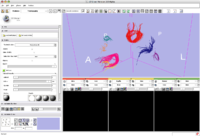
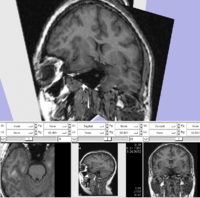
The SPL-PNL brain atlas loaded in Slicer3. Demonstrates model hierarchy and clipping.
Image from davisb on Monday, April, 27, 2007
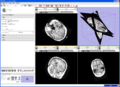
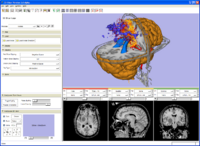
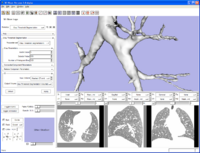
EMSegment screenshot---segmentation results and work-flow GUI
Image from magnotta on Monday, April, 16, 2007
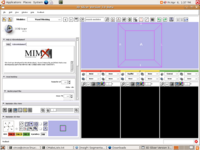
Mimx Logo in Slicer3 for VoxelMeshingModule
Image from blezek on Tuesday, April, 10, 2007 at 8:00AM
Python incorporated into Slicer.
Bringing up the Python console
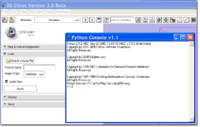
Now, do something interesting with it!
>>> # import MatPlotLib via pylab functions so we can refer to them directly >>> from pylab import * >>> imshow ( data[128,:,:] ) >>> # Instruct pylab to show our plots... hit return to continue entering commands. >>> show()
Voila!
Image from wjp on Thursday, March 22, 2007 at 10:00AM
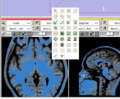
New GUI elements: Shows a magnified view in the GUI panel of an area around the mouse in any Slice Window.
Image from wjp on Thursday, March 22, 2007 at 10:00AM
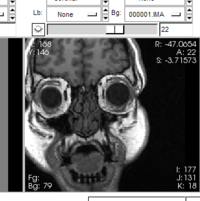
New GUI elements: Shows birds-eye-view of the scene relative to the outline of the 3D Viewer's window in the 'Manipulate 3D View' GUI panel.
Image from pieper on Tuesday, January 16, 2007 at 7:58PM
From Lauren, shows fiber tracts loaded from a file and with display properties controlled by the GUI.
Image from wjp on Friday, December 29, 2006 at 3:23PM
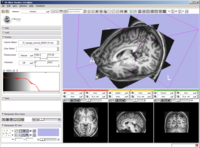 Module choose and navigation functionality integrated into the application toolbar to create more space for module GUIs on the side panel. Slice Controller widgets also updated to have more functionality available, a button to link and unlink their control, and an improved visual design.
Module choose and navigation functionality integrated into the application toolbar to create more space for module GUIs on the side panel. Slice Controller widgets also updated to have more functionality available, a button to link and unlink their control, and an improved visual design.
Image from wjp on Monday, November 28, 2006 at 6:40M
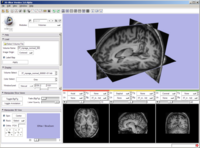 New 3D Slicer logo integrated with slicer GUI.
New 3D Slicer logo integrated with slicer GUI.
Image from pieper on Thursday, October 26, 2006 at 1:54M
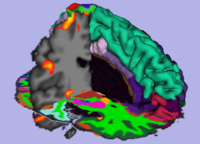 FreeSurfer structural data (cortical and subcortical segmentation) with fBIRN functional overlay, shown as part of Slicer3-based BIRN Query Atlas project.
FreeSurfer structural data (cortical and subcortical segmentation) with fBIRN functional overlay, shown as part of Slicer3-based BIRN Query Atlas project.
Image from pieper on Thursday, October 05, 2006 at 1:08PM
 Clipping with the slice plane is now supported, along with thresholded image display.
Clipping with the slice plane is now supported, along with thresholded image display.
Image from naucoin on Friday, September 08, 2006 at 12:14pm
The FreeSurfer colour lookup table has been added to the Slicer3 FreeSurfer Library. A curvature overlay file was loaded and displayed on bert/surf/lh.pial as loaded in via the Models module.
Image from naucoin on Friday, September 08, 2006 at 11:24am
The FreeSurfer scalar reader has been integrated as a Slicer 3 Library, and surfaces can be read via the Models module. Shot shows standard bert/surf/lh.pial plus a test script to load the annotation labels file.
Image from pieper on Tuesday, August 08, 2006 at 4:26PM
 A number of things have come together to allow the new prototype editor module:
A number of things have come together to allow the new prototype editor module:
- the label map layers are enabled
- lookup table support in the layer logic
- a new set of 2D UI widgets
- generalized painting infrastructure, which is based on requests and feedback from NA-MIC Core 3 users and other users of Slicer2
- paint brush has a 'threshold' option in which the label map only gets painted if the background intensity is in a specified range
- paint brush optionally won't overpaint existing voxels
- 'eye dropper' mode allows you to start painting with the label value where you first clicked (so you don't need to go to the UI to change the label color as often).
- all painting operations can happen on arbitrarily transformed volumes. This allows the painting plane to be aligned with the major axis of a structure like the hippocampus
- background layers are interpolated to current screen resolution (not limited to nearest neighbor sampling as in slicer2's draw mode)
- painting is performed using a circular brush with a diameter selected by the user. this makes it easier to follow the contours of curved biological structures.
- the cursor is turned off on mouse down and reappears on mouse up so you can see the exact pixels you are drawing (a circle around the cursor location indicates the mouse location)
A few limitations make this still a 'prototype' including:
- there's no convenient way to save your label maps volumes (except as dicom)
- the color map is currently random
- only label numbers 0-255 will show as distinct colors
- no hot keys for frequently used operations
- brush radius is in screen space units rather than RAS (means that the brush is effectively getting bigger and smaller as you zoom in and out)
- the paint algorithm is implemented inefficiently and can be slow when painting large numbers of pixels
- much of the code is written in tcl - new vtk classes are needed to implement the functionality more efficiently.
Image from pieper on Friday, July 28, 2006 at 9:04AM
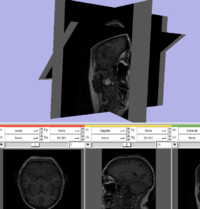 Test of making a non-orthogonal slice plane. This was created by manually setting the SliceToRAS matrix in the vtkMRMLSliceNode. GUI is not yet hooked up for oblique slices.
Test of making a non-orthogonal slice plane. This was created by manually setting the SliceToRAS matrix in the vtkMRMLSliceNode. GUI is not yet hooked up for oblique slices.
Image from pieper on Thursday, July 27, 2006 at 5:21PM
![]() Shows that a single pixel voxel in black is exactly bounded by a unit cube in RAS space. Also the blinking eye buttons for slice visiblility are hooked up and there is now a red-yellow-green color coding for the slice windows (carried over from slicer2).
Shows that a single pixel voxel in black is exactly bounded by a unit cube in RAS space. Also the blinking eye buttons for slice visiblility are hooked up and there is now a red-yellow-green color coding for the slice windows (carried over from slicer2).
Image from pieper on Thursday, July 13, 2006 at 4:49PM
 Test of interactive slice textured plane control. Not yet eneabled through GUI, but driven by a test script that you can source into the console.
Type Control-T to bring up interactor, then
Test of interactive slice textured plane control. Not yet eneabled through GUI, but driven by a test script that you can source into the console.
Type Control-T to bring up interactor, then
source Base/GUI/Testing/TestSliceModels.tcl SliceModelsAdd
This test does things the right way in that it creates model nodes for the slices and the updates of the planes is triggered by observers on the Slice Logic. But it does things the wrong way in that it creates the texture in an ad hoc way for now since we don't yet have a clean way to specify textures in the model display nodes. Also, the Viewer renders on every changes to the slice view, so as you pan/zoom/scroll the slices the redraws are jerky. Next step is to figure out how best to get this functionality integrated into the C++. Probably there should be a vtkSlicerSliceModelsLogic class that's managed by the vtkSlicerSliceGUI.
Image from pieper on Tuesday, July 11, 2006 at 3:39PM
 Example model read from vtk file and loaded in slicer3
Example model read from vtk file and loaded in slicer3
Image from pieper on Monday, July 03, 2006 at 3:35PM
 Test of the new logo widget for adding module-specific watermark logos directly in the 3d view. Uses a vtkLogoWidget from VTK cvs head. Kitware is working on cmake support to notify developers when a VTK cvs update is needed. Once that is done, the logo code will be added to slicer3 svn.
Test of the new logo widget for adding module-specific watermark logos directly in the 3d view. Uses a vtkLogoWidget from VTK cvs head. Kitware is working on cmake support to notify developers when a VTK cvs update is needed. Once that is done, the logo code will be added to slicer3 svn.
Image from pieper on Monday, June 26, 2006 at 10:38AM
 New annotations and key bindings. Middle mouse to pan, right mouse to zoom.
New annotations and key bindings. Middle mouse to pan, right mouse to zoom.
Image by pieper on Wednesday, June 21, 2006 at 1:01PM
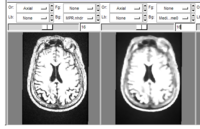 results of the command line median filter module
results of the command line median filter module
Image by millerjv on Tuesday, June 20, 2006 at 4:37PM
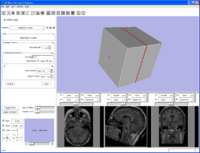
 {First command line module to export data, execute, and import data back into Slicer}
{First command line module to export data, execute, and import data back into Slicer}
Image by millerjv on Friday, June 16, 2006 at 11:15AM
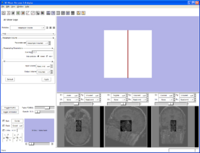
 {Command line module with an enumerated parameter as a radio button}
{Command line module with an enumerated parameter as a radio button}
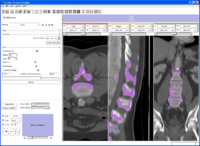
Image by millerjv on Wednesday, June 14, 2006 at 10:20AM
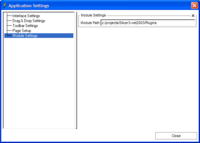 {Slicer applications settings panel showing the module search path}
{Slicer applications settings panel showing the module search path}
Image from pieper on Tuesday, June 13, 2006 at 8:23PM
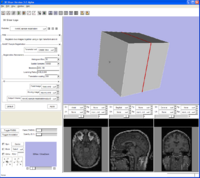 Another command line module example from Jim
Another command line module example from Jim
Image from pieper on Tuesday, June 13, 2006 at 8:20PM
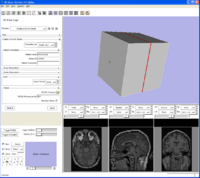 Example from Jim Miller showing the automatic GUI generation from XML collected from execution module commands
Example from Jim Miller showing the automatic GUI generation from XML collected from execution module commands
Image from pieper on Tuesday, June 13, 2006 at 8:18PM
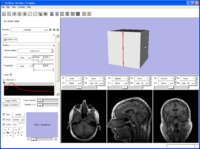 Example screen shot showing that there is a basic interface in place.
Example screen shot showing that there is a basic interface in place.
About the VisualBlog
The VisualBlog is meant to be an easy place to upload screenshot so that both developers and outside observes can track the progress of the project.
Upload Script
To simplify the process of uploading images and blog entries, you can use a helper script called wikimage which uploads the image and caption to this page. You must have a Tcl/Tk and curl installed on your machine to use wikimage.
NOTE: tested and working on Windows with Cygwin, needs debugging on Linux.
Example usage ./wikimage.tcl screenshot-of-my-new-feature-2006-06-14.png This shows my neat new slicer feature that I added in just minutes of pleasureable coding...
You will need to have a wiki account in order to do the upload. If you're wiki account isn't the same name as your local machine login, use the --user command line option to specify it.
Full usage:
usage: wikimage [options] (image_filename) (caption) (image_filename) is the file to upload (caption) is the rest of the command line [options] is one of the following: --gallery : wiki page where image link should be added --thumb-size : size of gallery thumbnail (default 200px) -u --user : wiki user name (default is env USER) -p --password : wiki login password --url : url of the wiki (default wiki.na-mic.org) -c --cookie-jar : file to save your cookies (default 'mycookies') --help : prints this message and exits
Of course after you've uploaded you can use regular wiki tools to edit and augment your caption.
Plans
We're looking into adding the wikimage command as menu item in slicer to further simplify, but for now it can be run on the command line.
Every month or so we can copy the contents of the page to an archive location so this page doesn't get too long.