2009 Summer Project Week Project Segmentation of Muscoskeletal Images
Key Investigators
- Stanford: Harish Doddi, Saikat Pal, Scott Delp
- Harvard: Ron Kikinis
- Steve Pieper, Isomics, Inc.
- Luis Ibanez, Kitware, Inc.
Objective
The aim of this project is to develop an automatic/semi-automatic methodology to convert whole body imaging datasets into three-dimensional models for neuromuscular biomechanics and finite element simulations.
Specific Aims
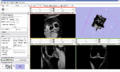
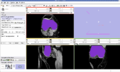
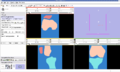
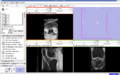
- Implement image-to-image registration and segmentation algorithms on knee MR datasets. Registration includes masking algorithms to isolate structures of interest (i.e. femur, patella, and tibia bones).
- Perform a parameter space exploration on masked MR datasets.
- Develop model-to-image registration technique to morph existing model atlas to specified MR image geometry.
Approach, Plan
We are working on understanding the capabilities of RegisterImage module in Slicer to apply to knee datasets. Currently we are conducting parameter exploration studies to evaluate the sensitivity of registered images to different input parameters associated with the algorithms. We are also developing a module to apply python ICP-based registration algorithms to directly morph a surface model to a target image geometry.
Our goals for the project week are:
- Perform and evaluate results from an extensive
parameter space exploration study of RegisterImages Batchmake module on knee dataset.
- Resolve issues in building Python modules from
slicer source code.
- Demonstrate proof of concept on registering an
existing atlas (.vtk, .stl) to a target image using Python ICP Registration module.
Progress