FSL-based segmentation pipeline
From NAMIC Wiki
Home < FSL-based segmentation pipeline
Abstract
This page describes the FSL-based EM segmentation. Page created by Aug, 09.
Segmentation pipeline
- Atlas construction phase:
- Chose one subject form the dataset as the target image;
- Using the Slicer3 module to perform histogram matching from all other subjects to the target;
- Affine register all subjects to the target using flirt;
- Manually skull strip the target;
- Apply the brain mask to all subjects to obtain brain matter;
- Perform bias correction on the brain matters using the binary within ITK Insight Applications.
- Affine register the brain matters of all subjects to that of the target using flirt, and save the transform as .mat;
- Subsequently, using Diffeomorphic Demons registration to register all subjects to the target and save the deformation field as .mha;
- Inverse the skull mask to obtain skull matter for all subjects;
- Apply the above obtained affine transformations and Demons deformations to deform their corresponding skull matter;
- Combine the brain matter and skull matter for each subject to obtain co-aligned brain images;
- Average the co-aligned images to generate the atlas;
- Apply KMS segmentation on all subjects, followed by manually correction of the segmentations;
- Collect all the segmentation results to generate tissue maps and the ECC masks.
- EM segmentation phase:
- Using the image of each subject as the target and the atlas as the source;
- For each subject, affine register its whole head image to the whole head atlas;
- Use the aligned brain mask from atlas to skull strip the subject image;
- Affine register the brain matter in the atlas to the masked brain matter of the subject, and save the transform as .mat;
- Subsequently, using Diffeomorphic Demons registration to register the brain matter in the atlas to that of the subject, and save the deformations as .mha;
- Apply the above obtained affine transformation and demons deformation to the tissue maps and ECC mask;
- Perform EM segmentation using the Slicer2/Slicer3 module.
Segmentation Results
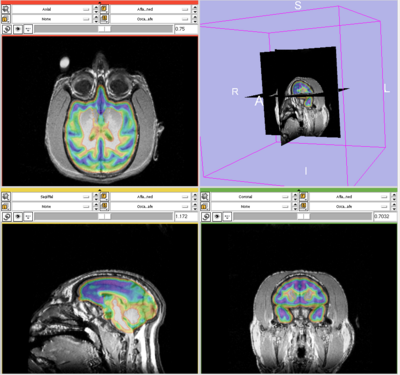
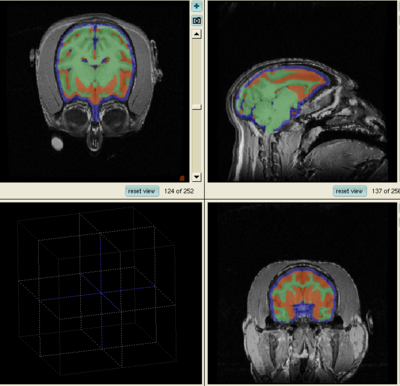
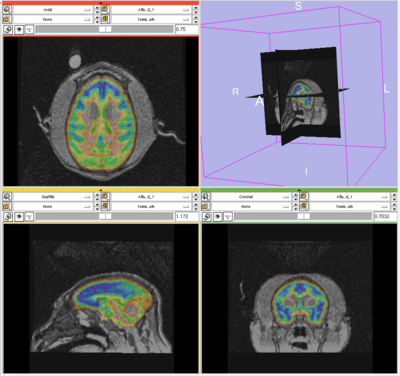
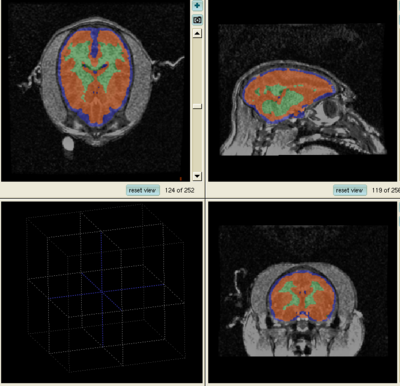
- The results for the best and worst segmentations are shown in the following. For each subject, the first figure shows the alignment of Gray matter atlas (these will be changed to checker board figures aligning target and the whole brain atlas soon) with the brains, and the second figure shows the segmentation results.
Best Segmentation: Oscar
Worst Segmentation: Tommy