Projects:dtistatisticsfibers
Statistics on DTI data
Back to NA-MIC Collaborations, Utah2 Algorithms, MIT Algorithms, UNC Algorithms
DTI Fiber Tract statistics
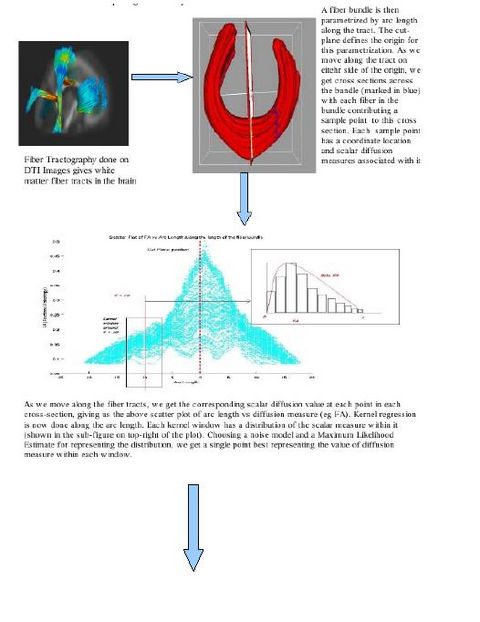
Following the complete pipeline using Casey Goodlett's work, we get the fiber files containing DTI data. (The pipeline includes unbiased non-rigid registration of a population, Fiber tractography applied to the average atlas, and finding individual subjects' DTI data by mapping the atlas geometry back to individual subjects.
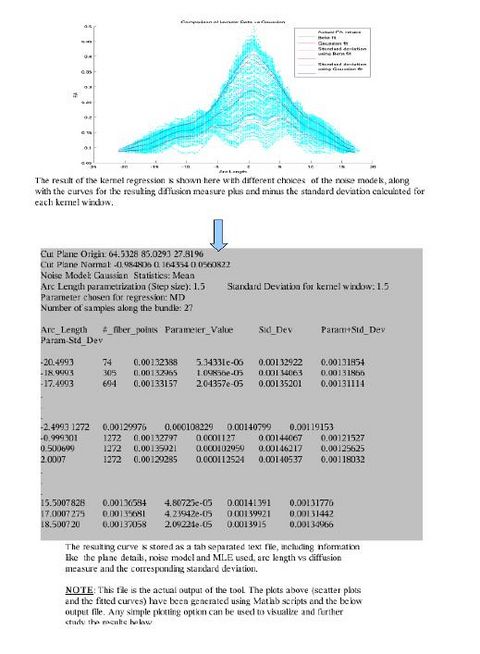
We are working on a command line tool which takes as input the fiber files generated by the above pipeline. The tool has options to perform kernel based regression on the DTI data (like FA, MD, FRO, AD, RD etc). Each kernel window assumes a noise model within the given cross section of the fiber and then chooses a statistic to represent the information in that window. Currently, the tool implements Gaussian and Beta (only for FA) noise models for data within fiber cross sections. The possible statistics that can be chosen are Mean, Mode and Quantiles. The tool also computes the standard deviations about the regressed curve. The output is saved as a file. All the results can be plotted using Matlab scripts.
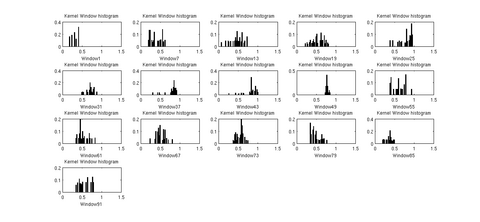
There are various visualization options available like viewing the data distribution histograms within cross sections (before and after applying the kernels), scatter plots to view the actual distribution of the DTI data, and more. These options will help in further analyzing the best noise models and the best representative statistics to explain the distribution of DTI data along the fiber tract.
The tool will replace the command line interface that Fiber Viewer tool currently provides to do similar (though very limited) tasks. It will allow us to do comparison of DTI data inter and intra population. We are currently testing the results of the tool on data from neonates, 1 year and 2 year old subjects. Casey's Functional Data Analysis module is also being used to analyze the results generated above.
Key Investigators
- Utah: Anuja Sharma, Guido Gerig
Objective
The aim is to understand probabilistic models that can account for the behavior of water diffusion in white matter tracts. The long term goal is to use this to understand the changes in white matter structure with age, gender or a specific disease.
Approach, Plan
Various tract-oriented scalar diffusion measures are treated as a continuous function of fiber arc-length. To analyze the trend along the fiber tract, a command line tool performs kernel regression on this data. The idea is to try out different noise models and maximum likelihood estimates within kernel windows, such that they best represent the data and are robust to noise and Partial Volume effect.
Casey Goodlett's functional data analysis pipeline is then applied to this data. Here, multivariate hypothesis test is used to test for differences between populations.
Our plan for the project week is to try out the complete pipeline for a few datasets and see if we get statistically significant results. We also plan to work on the possibility of integrating this module into the Slicer environment.
Progress
The first version of the command line tool is ready. It provides the flexibility to choose the scalar diffusion measure to be tested; a choice between Gaussian and Beta noise models and Mean, median or mode as MLE. It also incorporates several visualization options which help in analyzing the best noise models and the best representative statistics to explain the distribution of DTI data along the fiber tract.
This week we figured out all interoperability issues which need to fixed to enable the tool's integration into the Slicer. We discussed with other groups working on DTI and discussed common data structures and file formats for easy interfacing between tools. As a result, we will be moving our files to the VTK format in the near future.
Another discussion with Maddah pointed out ways to minimize the need for manual intervention in our tool. Moreover, we would be collaborating with the University of Iowa in providing tools to process datasets related to Huntington's disease. We might also migrate another medical imaging software Fiber Viewer to Qt4 to enable integration into the Slicer environment.
During this week we also tried running the pipeline on several datasets. We are in the process of analyzing the results now.
Progress - revised
The command line tool is ready for upload into the NITRC. The features available in the tool currently, its use and input / output formats and other relevant details are provided in the first draft of the documentation (PDF). The tool is still a work in progress. More features will be added to it, specially the option to plot and visualize the results.