Events:DTI Tractography Challenge MICCAI 2012
Note: Page under construction.
- Submission under consideration.

|

|

|
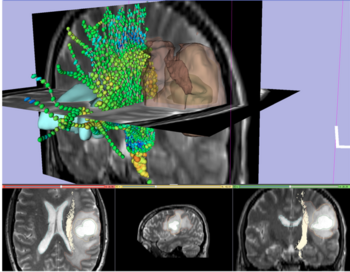
DTI Tractography for Neurosurgical Planning: A Grand Challenge
The goal of the DTI Tractography Challenge initiative is to provide neurosurgeons with an overview of cuting-edge tractography techniques developed in research for pre-surgical planning, and to give computer scientists and clinical researchers the opportunity to evaluate the performances of their tractography algorithms in a neurosurgical context, in the absence of ground truth. By bridging the gap between algorithm development and clinical use, this initiative aims at accelerating the transfer of the latest DTI technology to the neurosurgeons. The 1st edition of the MICCAI DTI Tractography Challenge was held as part of the 14th International Conference on Medical Image Computing and Computer Assisted Intervention MICCAI 2011 in Toronto, Canada.
Tentative Agenda (under construction)
- 8:30-10:00: Start of the on-site Challenge on neurosurgical cases
- 10:00-10:05: Welcoming remarks and Introduction
- 10:15-10:30: DTI Tractography validation challenge, introduction
- 10:30-10:45: DTI Tractography in the clinics
- 10:45-11:00: Presentation of the DTI challenge review criteria
- 11:00-11:15: Coffee-Break
- 11:15-12:15: Tractography Session: Presentation of tractography algorithms
- 12:15-13:30: Lunch Break
- 13:30: End of the Grand Challenge on neurosurgical cases
- 13:30-14:30: Clinical case review: presentation of the Grand Challenge cases by contestants
- 14:30-15:15: Keynote Speaker Lecture
- 15:15-16:15: Coffee Break and closed-review session
- 16:15-17:00: Presentation of the outcomes of the review
- 17:00-17:30: Announcement of the results and discussion
Workshop Datasets
The workshop datasets consist of anonymized neurosurgical cases of brain tumor patients, and repeated volunteer scans from two healthy subjects. Each dataset includes Anatomical Images (T1,T2), a Diffusion Weighted Imaging (DWI) volume, and a Diffusion Tensor Imaging (DTI) volume. Each clinical dataset includes a segmentation of the tumor. Data are in the ITK-readable Nrrd file format, which consists of an ASCII header file and a separate uncompressed raw image datafile.