Projects:ModelingFunctionalActivationPatterns
Back to NA-MIC Collaborations, MIT Algorithms,
Modeling Functional Activation Patterns
For a given cognitive task such as language processing, the location of corresponding functional regions in the brain may vary across subjects relative to anatomy. We present a probabilistic generative model that accounts for such variability as observed in functional magnetic resonance imaging (fMRI) data. We relate our approach to sparse coding that estimates a basis consisting of functional regions in the brain. Individual fMRI data is represented as a weighted sum of these functional regions that undergo deformations. We demonstrate the proposed method on a language fMRI study. Our method identified activation regions that agree with known literature on language processing and established correspondences among activation regions across subjects, producing more robust group-level effects than anatomical alignment alone.
Description
We propose a novel way to characterize functional variability that combines insights from prior work. We model each subject's activation map as a weighted sum of group-level functional activation parcels that undergo a subject-specific deformation. Similar to Xu et al. [1], we define a hierarchical generative model, but instead of using a Gaussian mixture model to represent shapes, we represent each parcel as an image, which allows for complex shapes. By representing each subject's activation in terms of group-level parcels, our model maintains parcel correspondences across subjects, similar to [2]. Next, we assume that the template regions can deform to account for functional variability. This involves using groupwise registration similar to [3] that is guided by estimated group-level functional activation regions. We perform inference within the proposed model using an algorithm similar to expectation-maximization (EM) and illustrate our method on the language system, which is known to have significant functional variability [4].
Experiments
We train our model on an fMRI study of 82 subjects reading sentences and pronounceable non-words [4]. First, we apply the standard fMRI general linear model for the sentences vs. non-words contrast. We apply our model to subjects' t-statistic maps thresholded at p-value=0.01. These images are pre-aligned using the subjects' anatomical MRI scans via affine registration.
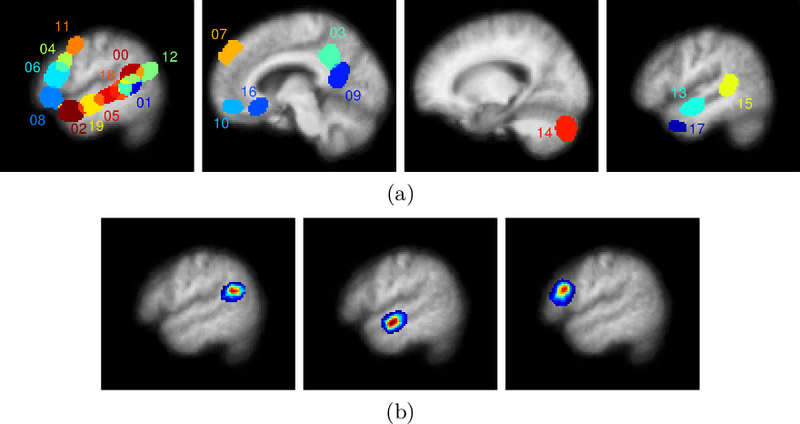
Fig. 1(a) shows the spatial support of the final learned dictionary elements on four slices. Fig. 1(b) illustrates some of the dictionary elements extracted by the algorithm. The dictionary elements include regions previously reported as indicative of lexical and structural language processing [4], namely portions of the temporal lobes, the right cerebellum, and the left frontal lobe. There are also dictionary elements corresponding to the medial prefrontal cortex, the posterior cingulate, and the precuneus.
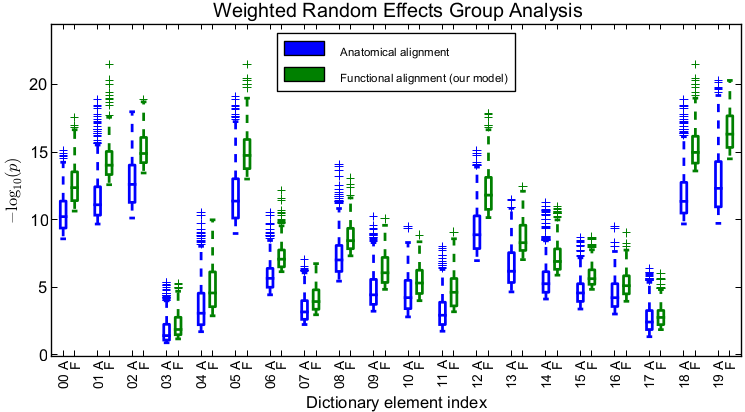
To evaluate the quality of the estimated alignment, we apply the estimated deformation to held-out time course data for each subject and perform standard weighted random effects analysis. We then look at significance values within the support of each dictionary element. Importantly, for drawing conclusions on the group-level parcels defined by the estimated dictionary elements, within each parcel, it is the peak and regions around the peak that are of interest rather than the full support of the dictionary element. Thus, to quantify the advantage of our method, within each dictionary element, we compare the top 25% highest significance values for our method versus those of anatomical alignment (Fig. 2). We observe that accounting for functional variability via deformations results in substantially higher peak significance values within the estimated group-level parcels, suggesting better overlap of these functional activation regions across subjects. On average, our method improves the significance of group analysis by roughly an order of magnitude when evaluating the top 25% significance values. Even if we look at the top 50% of significance values in each dictionary element, the results remain similar to those in Fig. 2.
Conclusion
We developed a model that accounts for spatial variability of functional activation regions in the brain via deformations of weighted dictionary elements. Learning model parameters and estimating deformations yield correspondences of functional activation regions in the brain across subjects. We demonstrate our model in a language fMRI study, which contains substantial variability. We plan to validate the detected parcels using data from different fMRI language experiments.
Literature
[1] L. Xu, T.D. Johnson, T.E. Nichols, and D.E. Nee. "Modeling inter-subject variability in fMRI activation location: a bayesian hierarchical spatial model". Biometrics 65(4), 1041–1051, 2009.
[2] B. Thirion, P. Pinel, A. Tucholka, A. Roche, P. Ciuciu, J.F. Mangin, J.B. Poline. "Structural analysis of fMRI data revisited: improving the sensitivity and reliability of fMRI group studies". IEEE Transactions in Medical Imaging 26(9), 1256–1269, 2007.
[3] M.R. Sabuncu, B.D. Singer, B. Conroy, R.E. Bryan, P.J. Ramadge, J.V. Haxby. "Function-based intersubject alignment of human cortical anatomy". Cerebral Cortex 20(1), 130–140, 2010.
[4] E. Fedorenko, P.J. Hsieh, A. Nieto-Castanon, S. Whitfield-Gabrieli, N. Kanwisher. "New method for fMRI investigations of language: Defining ROIs functionally in individual subjects". Neurophysiology 104, 1177–1194, 2010.
Key Investigators
MIT: George H. Chen, Evelina G. Fedorenko, Nancy G. Kanwisher, and Polina Golland
Publications
NA-MIC Publications Database on Modeling Functional Activation Patterns