Visualization and quantification of peri-contusional white matter bundles in traumatic brain injury using diffusion tensor imaging
From NAMIC Wiki
Home < Visualization and quantification of peri-contusional white matter bundles in traumatic brain injury using diffusion tensor imaging
Key Investigators
- UCLA: Andrei Irimia, Micah C. Chambers, Ron Kikinis, Paul M. Vespa, Jack Van Horn
Objective
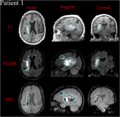
The longitudinal fate of peri-contusional white matter (WM) is of appreciable clinical interest in neurotraumatology due to wide variability of outcome scenarios, ranging from tissue survival to atrophy to permanent tissue loss. The ability to predict WM fate in acute TBI patients would be extremely helpful from a clinical standpoint for the purpose of surgical planning and for the personalized formulation of rehabilitation treatments. This project is concerned with using 3D Slicer to visualize and quantify peri-contusional WM bundles extracted from TBI volumes acquired via DTI.
Approach, Plan
- we intend to perform an exploratory multimodal analysis of structural effects upon WM due to intracranial hemorrhage (ICH) in three selected clinical cases of progressive severity
- tissue classification will be performed following the established methods of Irimia et al., including multimodal segmentation followed by model rendering in 3D Slicer
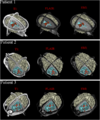
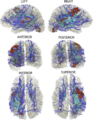
- peri-lesional fiber bundles and the cortico-spinal tract will be reconstructed using DTI deterministic tractography using 3D Slicer
- tract streamline visualization will be performed to identify tracts within a 4 mm proximity to each lesion
- fiber deformation and bending around lesions will be characterized and quantified
Progress
- we have performed an exploratory multimodal analysis of structural effects upon white matter due to intracranial hemorrhage
- multimodal segmentation was performed in Slicer to identify peri-lesional areas
- the putative boundary between pathology and healthy-appearing tissues was identified subsequent to the multimodal segmentation
- an algorithm was implemented to identify peri-lesional fiber bundles, i.e. fibers which passed within ~4 mm of each intracranial lesion
- three-dimensional models of pathology were generated in 3D Slicer in order to simultaneously visualize the white matter structure as well as the anatomical pathology related to the intracranial bleeding
References
- Andrei Irimia and John D. van Horn (2012) The structural, connectomic and network covariance of the human brain NeuroImage (accepted) [1]
- Andrei Irimia, Bo Wang, Stephen R. Aylward, Marcel W. Prastawa, Danielle F. Pace, Marc Niethammer, Guido Gerig, David A. Hovda, Ron Kikinis, Paul M. Vespa and John D. van Horn (2012) Multimodal neuroimaging of structural pathology and connectomics in traumatic brain injury: toward personalized outcome prediction Neuroimage: Clinical volume 1, pages 1-17 [2]
- John D. van Horn, Andrei Irimia, Carinna M. Torgerson, Micah C. Chambers, Arthur W. Toga (2012) Mapping connectivity damage in the case of Phineas Gage. Plos ONE [3]
- Andrei Irimia, Micah C. Chambers, Carinna M. Torgerson and John D. van Horn (2012) Circular representation of human cortical networks for subject and population-level connectomic visualization NeuroImage volume 60, pages 1340-1351
- Andrei Irimia, Micah C Chambers, Carinna M Torgerson, Maria Filippou, David A Hovda, Jeffry R Alger, Guido Gerig, Arthur W Toga, Paul M Vespa, Ron Kikinis, John D Van Horn (2012) Patient-tailored connectomics visualization for the assessment of white matter atrophy in traumatic brain injury. Frontiers in Neurology [4]
- Andrei Irimia, Micah C. Chambers, Jeffry R. Alger, Maria Filippou, Marcel W. Prastawa, Bo Wang, David A. Hovda, Guido Gerig, Arthur W. Toga, Ron Kikinis, Paul M. Vespa, John D. van Horn (2011) Comparison of acute and chronic traumatic brain injury using semi-automatic multimodal segmentation of MR volumes. Journal of Neurotrauma [5]