Projects:DiffeoMixedEffects
Back to Utah 2 Algorithms
Ongoing Work
Diffeomorphic Shape Trajectories for Improved Longitudinal Segmentation and Statistics
Longitudinal image data has several sources of variability. First, there is inherent biological variability, both within a subject changing over time and also between subjects in a population. The goal of longitudinal analysis is to quantify this variability and make inferences about changes over time in a population. However, longitudinal imaging data also include unwanted sources of variability, such as noise in image acquisition, segmentation and registration errors, and human expert judgment, among others. These extraneous errors tend to dampen statistical power, especially when trying to distinguish between trajectories of two different populations, e.g., healthy and diseased.
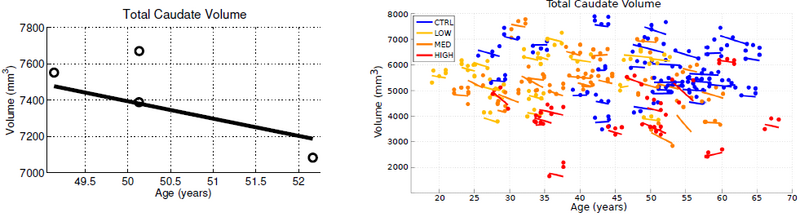
We study subcortical change associated with Huntington’s disease (HD), leveraging the longitudinal study PREDICT-HD. The longitudinal database consists of 65 female subjects: 23 controls (CTRL), 14 (LOW), 15 (MED), and 13 (HIGH). The LOW / MED / HIGH categories represent probability of onset of manifesting signs of HD. All subjects have had at least 3 MR images acquired approximately one year apart, with many subjects undergoing multiple scans per visit. Six subcortical pairs (caudate, putamen, hippocampus, thalamus, acumben, and pallidus) were segmented from each image and manually verified and cleaned.
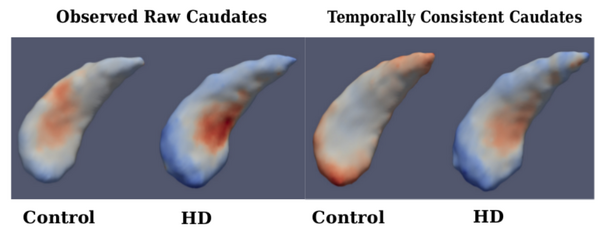
The quality of each segmentation varies considerably for each time point, even when scans are obtained on the same day from the same scanner, as individual single-subject segmentation is prone to errors related to variability of imaging, image calibration, human expert judgment, and limited robustness of segmentation algorithms. While the segmentation quality is not easily assessed by viewing the 3D anatomical surfaces, the temporal inconsistency becomes clear by investigating volume extracted from the shapes. The left side of Fig. 1 shows the variability in segmentation, illustrated by the temporal inconsistency of observed caudate volume, motivating the need for temporally consistent segmentations which properly account for correlated longitudinal data.
Continuous models of shape trajectory are estimated for each subject, resulting in personalized and temporally consistent anatomical evolution. Model estimation does not require point correspondence, facilitating the inclusion of all subcortical shapes simultaneously without imposing any topological constraints. Each subject’s personalized model allows us to generate shapes at any instant in time, from which desired shape properties, such as volume, can be extracted. We can therefore obtain a continuous evolution of volume for all subcortical structures without any explicit modeling of volume. Fig. 1 shows caudate volume extracted from each subject’s continuous shape model, demonstrating the flexibility of the shape model to capture both linear and non-linear volume trends with no prior assumption or constraint on linearity. Though we only display caudate volume here, recall that each model is estimated by leveraging all shape data simultaneously (Fig. 2), which respects shape boundaries and locations, incorporating important geometric relationships between shapes.

Literature
[1] Muralidharan, P., Fishbaugh, J., Johnson, H., Durrleman, S., Paulsen, J., Gerig, G., Fletcher, P.T. Diffeomorphic shape trajectories for improved longitudinal segmentation and statistics. Proc. of Medical Image Computing and Computer Assisted Intervention (MICCAI '14). (2014).
Key Investigators
- Utah: Prasanna Muralidharan, James Fishbaugh, P.T. Fletcher, Guido Gerig
- INRIA/ICM, Pitie Salpetriere Hospital: Stanley Durrleman
- Department of Psychiatry, Carver College of Medicine, University of Iowa: Jane Paulsen, Hans Johnson