T1 mapping for variable flip angle
From NAMIC Wiki
Home < T1 mapping for variable flip angle
Key Investigators
- MGH: Xiao Da, Artem Mamonov, Jayashree Kalpathy-Cramer
- BWH: Andriy Fedorov
Project Description
Objective
- Estimate effective T1 from multi-spectral FLASH MRI scans with different flip angles
- Implement T1 mapping algorithm as a Slicer extension.
Approach, Plan
- Create a Slicer module for T1 mapping for multiple flip angles using Python script
- Run Python script and Freesurfer code on QIBA T1 phantom
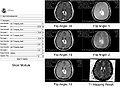
- Test the Slicer module on MGH Brain Tumor MR Data with multiple flip angles
- Go through Slicer tutorials and developer forum to learn how to implement Slicer extensions.
- Go over VTK documentation to learn how it works.
Progress
- Compared the results using Python with Freesurfer and ground truth of T1 for QIBA data
- T1 mapping results on QIBA data using Slicer+Python are almost identical to the results using Freesurfer and comparable with the ground truth.
- Implemented T1 mapping as a Slicer extension using python. The extension has GUI that as an input takes 6 DICOM images with corresponding flip angles and repetition time. The extension computes T1s and stores them in a new VTK volume node.