Difference between revisions of "2008 Engineering review at UNC"
From NAMIC Wiki
| (33 intermediate revisions by 3 users not shown) | |||
| Line 5: | Line 5: | ||
'''[[2008_Engineering_review_at_UNC:Summary | SUMMARY AND ACTION ITEMS]]''' | '''[[2008_Engineering_review_at_UNC:Summary | SUMMARY AND ACTION ITEMS]]''' | ||
| + | |||
'''[[2008_Engineering_review_at_UNC:FollowUp | ACTION UPDATE]]''' | '''[[2008_Engineering_review_at_UNC:FollowUp | ACTION UPDATE]]''' | ||
| − | ===Suggested | + | ===Minutes=== |
| + | |||
| + | * General issues: | ||
| + | ** Source / Module handling | ||
| + | *** Current idea: Source updated within UNC NeuroLib - Releases at NITRIC | ||
| + | *** Steve: presentation of ideas to load/unload Slicer extensions | ||
| + | *** It would be important to have easy access to both Nightly and Release versions | ||
| + | *** NAMIC Wiki for roadmap description | ||
| + | *** Slicer Wiki for modules, NITRC links to this documentation | ||
| + | *** Install compilation of Modules for upload/download | ||
| + | |||
| + | ** Population handling | ||
| + | *** XNAT browse within Slicer | ||
| + | *** createx xcat => included in Batchmake for running on population | ||
| + | *** This should be used for the current development of ARCTIC population phase | ||
| + | |||
| + | ** Download of a module | ||
| + | *** Wizard selection | ||
| + | *** SLICER_MODULE_PATHS => for executables & BatchMake bmm files | ||
| + | |||
| + | ===Suggested Hotels=== | ||
| + | |||
| + | Nice for walking to campus & Franklin Street, but rather expensive: | ||
| + | * [http://www.carolinainn.com/ Carolina Inn], 211 Pittsboro Street, Chapel Hill, NC | ||
| + | * [http://www.franklinhotelnc.com/ Franklin Hotel], 311 W Franklin St, Chapel Hill, NC | ||
| + | |||
| + | Best suggestion (needs taxi to campus/UNC Hospitals & Franklin Street): | ||
| + | * [http://www.courtyardchapelhill.com/ Courtyard Marriott], 100 Marriott Way, Chapel Hill, NC | ||
===Engineering Attendees (projected)=== | ===Engineering Attendees (projected)=== | ||
Will Schroeder, Steve Pieper, (possibly) Jim Miller, Julien Jomier | Will Schroeder, Steve Pieper, (possibly) Jim Miller, Julien Jomier | ||
| − | ===Place=== | + | ===Place === |
| + | |||
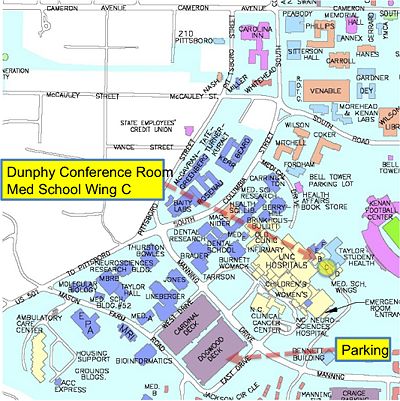
| + | * University of North Carolin, Chapel Hill | ||
| + | * Dunphy Conference Room (see also map) | ||
| + | * 3rd Floor Medical School Wing C (next to Neuro Image Research and Analysis Lab) | ||
| + | * There is very limited parking on campus for visitors, so car rental is not necessarily encouraged, but nearest parking is in Dogwood UNC Hospital Patient & Visitor Parking House (see map) | ||
| + | |||
| + | * Transportation from Airport | ||
| + | ** Taxi ride to UNC Hospitals: 30-35 USD, use map or call Martin (919-260-6674) to get to the conference room | ||
| + | |||
| + | [[Image:DunphyConfRoomLocation.jpg|400px]] | ||
===Proposed discussion items=== | ===Proposed discussion items=== | ||
| Line 19: | Line 57: | ||
===Schedule=== | ===Schedule=== | ||
| + | Tentative schedule: | ||
| + | |||
| + | Monday: noon - 5pm | ||
| + | * Noon - 1 pm: Arrival, Lunch and Meet of the whole group | ||
| + | * 1pm - 2pm: UNC cortical thickness pipeline and related issues (regional analysis) - Vachet, Mathieu, Styner | ||
| + | * 2pm - 3pm: UNC cortical thickness pipeline for local analysis - Vachet, Styner, Mathieu | ||
| + | * 3pm - 4pm: UNC cortical/surface correspondence incorporating DTI - Oguz, Styner, Vachet | ||
| + | * 4pm - 5pm: Make up time and wrap up for the day | ||
| + | |||
| + | * 7pm: Dinner on Franklin Street: Restaurant TBA | ||
| + | |||
| + | Tuesday: 9am - 1pm | ||
| + | * 9am - 10am: UNC shape analysis framework, including MANCOVA based statistical analysis - Styner | ||
| + | * 10am - 10:45am: DTI prep tool - Liu, Styner | ||
| + | * 10:45am - 11:30: DTI voxel based analysis - Liu, Styner | ||
| + | * 11:30am - noon: Mouse brain analysis pipeline and Slicer 3 - Lee, Styner | ||
| + | * noon - 1pm: Make up time and wrap up | ||
| + | |||
| + | * General topics: BatchMake, Grid computing, Population tools in Slicer, NITRC and Slicer, Atlases, MRML File Output, Workflow Wizards | ||
| + | |||
| + | * Saving Parameter sets (save MRML parameters or could be BatchMake config file) | ||
| + | |||
| + | Catering: | ||
| + | MEDITERRANEAN DELI [http://www.mediterraneandeli.com/Catering_Menu/catering_menu.html Menu] | ||
===Attendees=== | ===Attendees=== | ||
| Line 30: | Line 92: | ||
* Steve Pieper | * Steve Pieper | ||
* Jim Miller | * Jim Miller | ||
| + | * Marc Niethammer (possibly) | ||
| + | |||
| + | ===Notes=== | ||
| + | |||
| + | Introductory Comments | ||
| + | * Intention is to replace freesurfer with a open platformbetter | ||
| + | * Making the source code available | ||
| + | ** talked at length about Slicer module availability including nightly versions of them | ||
| + | * Discussed the meaning of "Regional" analysis (as compared to "local" analysis). | ||
| + | * Began discussing the integration of BatchMake into Slicer; this discussion made up a good part of the trip. | ||
| + | |||
| + | * Regional Cortical Thickness Pipeline ARCTIC | ||
| + | ** spoke at length about BatchMake integration including xcat (which is already supported) | ||
| + | |||
| + | * Floated the idea of a MRML browser. On later reflection we wondered what we could do to modify the current Slicer to avoid supporting multiple apps. | ||
| + | |||
| + | * The need for compiling VTK in offscreen mode came up. We are interested in creating thumbnails and embedding them in MRML, etc. We will determine the best way to do this. | ||
| + | |||
| + | * SLICER_MODULE_PATHS needs to be added for BatchMake support. We are automating the setup of the workflow | ||
| + | |||
| + | * Long discussion about creating output directories and output parameter sets. | ||
| + | |||
| + | * As far as SQUISH and GUI testing, we may have to work with the SQUISH guys to embed the VTK's image difference filter into their testing process. | ||
| + | |||
| + | * Shape analysis | ||
| + | ** particular attention on XNAT, BatchMake and relationship to the current pipeline | ||
| + | ** the addition of XNAT will require rewriting much of the current pipeline, part of the complexity is due to the need to extract metadata and actual data from XNAT, and then introduce it into the pipeline. | ||
| + | * DTIPrep GUI and pipeline was discussed | ||
| + | * Reviewed the mouse brain processing | ||
| + | * Talked about BatchMake having the ability to generate a GUI | ||
| + | * Saving Slicer template files that BatchMake can fill in for later processing | ||
| + | |||
| + | Slicer Items | ||
| + | * Scene matrix dialog | ||
| + | ** parse directory tree or use config file | ||
| + | ** show matrix of subjects and QC steps for easy loading | ||
| + | ** ideally with screen captures of mrml scene | ||
| + | * screen grabs of mrml scenes at load and or save time (or on demand during processing) saved in data dir | ||
| + | * Model intersection visible in model display gui | ||
| + | * BatchMake build as part of genlib | ||
| + | * Slicer3_Module_Paths variable exported to environment for BatchMake to inherit | ||
| + | * UNC DTI QC module uses in TBI | ||
| + | * "Decorated" BatchMake to act as Slicer CLI by generating --xml to build gui for configuring script | ||
| + | * "Open As Template..." for MRML scene file | ||
| + | ** all Storable nodes listed in dialog | ||
| + | ** user can accept default or browse to files to override | ||
Latest revision as of 13:50, 22 December 2008
Home < 2008 Engineering review at UNCDate: December 15-16, 2008.
Arrival: Morning of Monday, December 15. Leaving: Afternoon on Tuesday, December 16.
Contents
Minutes
- General issues:
- Source / Module handling
- Current idea: Source updated within UNC NeuroLib - Releases at NITRIC
- Steve: presentation of ideas to load/unload Slicer extensions
- It would be important to have easy access to both Nightly and Release versions
- NAMIC Wiki for roadmap description
- Slicer Wiki for modules, NITRC links to this documentation
- Install compilation of Modules for upload/download
- Source / Module handling
- Population handling
- XNAT browse within Slicer
- createx xcat => included in Batchmake for running on population
- This should be used for the current development of ARCTIC population phase
- Population handling
- Download of a module
- Wizard selection
- SLICER_MODULE_PATHS => for executables & BatchMake bmm files
- Download of a module
Suggested Hotels
Nice for walking to campus & Franklin Street, but rather expensive:
- Carolina Inn, 211 Pittsboro Street, Chapel Hill, NC
- Franklin Hotel, 311 W Franklin St, Chapel Hill, NC
Best suggestion (needs taxi to campus/UNC Hospitals & Franklin Street):
- Courtyard Marriott, 100 Marriott Way, Chapel Hill, NC
Engineering Attendees (projected)
Will Schroeder, Steve Pieper, (possibly) Jim Miller, Julien Jomier
Place
- University of North Carolin, Chapel Hill
- Dunphy Conference Room (see also map)
- 3rd Floor Medical School Wing C (next to Neuro Image Research and Analysis Lab)
- There is very limited parking on campus for visitors, so car rental is not necessarily encouraged, but nearest parking is in Dogwood UNC Hospital Patient & Visitor Parking House (see map)
- Transportation from Airport
- Taxi ride to UNC Hospitals: 30-35 USD, use map or call Martin (919-260-6674) to get to the conference room
Proposed discussion items
Schedule
Tentative schedule:
Monday: noon - 5pm
- Noon - 1 pm: Arrival, Lunch and Meet of the whole group
- 1pm - 2pm: UNC cortical thickness pipeline and related issues (regional analysis) - Vachet, Mathieu, Styner
- 2pm - 3pm: UNC cortical thickness pipeline for local analysis - Vachet, Styner, Mathieu
- 3pm - 4pm: UNC cortical/surface correspondence incorporating DTI - Oguz, Styner, Vachet
- 4pm - 5pm: Make up time and wrap up for the day
- 7pm: Dinner on Franklin Street: Restaurant TBA
Tuesday: 9am - 1pm
- 9am - 10am: UNC shape analysis framework, including MANCOVA based statistical analysis - Styner
- 10am - 10:45am: DTI prep tool - Liu, Styner
- 10:45am - 11:30: DTI voxel based analysis - Liu, Styner
- 11:30am - noon: Mouse brain analysis pipeline and Slicer 3 - Lee, Styner
- noon - 1pm: Make up time and wrap up
- General topics: BatchMake, Grid computing, Population tools in Slicer, NITRC and Slicer, Atlases, MRML File Output, Workflow Wizards
- Saving Parameter sets (save MRML parameters or could be BatchMake config file)
Catering: MEDITERRANEAN DELI Menu
Attendees
- Martin Styner
- Clement Vachet
- Cedric Mathieu
- Ipek Oguz
- Zhexing Liu
- Julien Jomier
- Will Schroeder
- Steve Pieper
- Jim Miller
- Marc Niethammer (possibly)
Notes
Introductory Comments
- Intention is to replace freesurfer with a open platformbetter
- Making the source code available
- talked at length about Slicer module availability including nightly versions of them
- Discussed the meaning of "Regional" analysis (as compared to "local" analysis).
- Began discussing the integration of BatchMake into Slicer; this discussion made up a good part of the trip.
- Regional Cortical Thickness Pipeline ARCTIC
- spoke at length about BatchMake integration including xcat (which is already supported)
- Floated the idea of a MRML browser. On later reflection we wondered what we could do to modify the current Slicer to avoid supporting multiple apps.
- The need for compiling VTK in offscreen mode came up. We are interested in creating thumbnails and embedding them in MRML, etc. We will determine the best way to do this.
- SLICER_MODULE_PATHS needs to be added for BatchMake support. We are automating the setup of the workflow
- Long discussion about creating output directories and output parameter sets.
- As far as SQUISH and GUI testing, we may have to work with the SQUISH guys to embed the VTK's image difference filter into their testing process.
- Shape analysis
- particular attention on XNAT, BatchMake and relationship to the current pipeline
- the addition of XNAT will require rewriting much of the current pipeline, part of the complexity is due to the need to extract metadata and actual data from XNAT, and then introduce it into the pipeline.
- DTIPrep GUI and pipeline was discussed
- Reviewed the mouse brain processing
- Talked about BatchMake having the ability to generate a GUI
- Saving Slicer template files that BatchMake can fill in for later processing
Slicer Items
- Scene matrix dialog
- parse directory tree or use config file
- show matrix of subjects and QC steps for easy loading
- ideally with screen captures of mrml scene
- screen grabs of mrml scenes at load and or save time (or on demand during processing) saved in data dir
- Model intersection visible in model display gui
- BatchMake build as part of genlib
- Slicer3_Module_Paths variable exported to environment for BatchMake to inherit
- UNC DTI QC module uses in TBI
- "Decorated" BatchMake to act as Slicer CLI by generating --xml to build gui for configuring script
- "Open As Template..." for MRML scene file
- all Storable nodes listed in dialog
- user can accept default or browse to files to override