Difference between revisions of "2008 Winter Project Week:Lesions"
Hjbockholt (talk | contribs) |
|||
| (21 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:NAMIC-SLC.jpg|thumb|320px|Return to [[2008_Winter_Project_Week]] ]] | |[[Image:NAMIC-SLC.jpg|thumb|320px|Return to [[2008_Winter_Project_Week]] ]] | ||
| − | |valign="top"|[[Image: | + | |valign="top"|[[Image:Lupus.png|320px]] |
|} | |} | ||
| Line 8: | Line 8: | ||
* MIND: Jeremy Bockolt, Mark Scully | * MIND: Jeremy Bockolt, Mark Scully | ||
* BWH: Sonia Pujol | * BWH: Sonia Pujol | ||
| + | * Utah: Marcel Prastawa | ||
| + | * Iowa: Vincent Magnotta | ||
* Kitware: Brad Davis | * Kitware: Brad Davis | ||
| Line 15: | Line 17: | ||
<h1>Objective</h1> | <h1>Objective</h1> | ||
| − | + | Our objective is to make progress on the roadmap initiative of creating an end to end solution that provides a tutorial for NA-MIC kit users to perform lesion analyses in Slicer3. | |
</div> | </div> | ||
| Line 22: | Line 24: | ||
<h1>Approach, Plan </h1> | <h1>Approach, Plan </h1> | ||
| − | + | For this week, we will attend and participate in the following training relevant to this DBP activities: | |
| + | *EM Segmenter User Group | ||
| + | *Plugins for Slicer3 | ||
| + | *DBP Engineers Lunch | ||
| + | *Registration Breakout | ||
| + | *Batchmake | ||
| + | |||
| + | Fully analyze 2 lupus subjects and 2 matched healthy normal volunteer data-sets collected: | ||
| + | *EM Segment | ||
| + | *BRAINS | ||
| + | *Prastawa/Gerig Lesion analysis method | ||
| + | |||
| + | We see our roadmap initiative breaking into the following stages: | ||
| + | |||
| + | a) Criteria and definition of what does an expert define and perceive as a | ||
| + | lesion (like rules on multi-contrast MRI), creation of a | ||
| + | document/catalogue that guides manual segmentation but also algorithm | ||
| + | developers. | ||
| + | |||
| + | b) Specification of advanced software for user-guided | ||
| + | efficient segmentation of lesions (e.g. 3D level set evolution etc., | ||
| + | definition of MRI contrast to use to do that, etc.). | ||
| + | |||
| + | c) Towards approaches towards fully automatic lesion segmentation using | ||
| + | multi-contrast MRI., | ||
| + | |||
| + | d) Validateion/test framework for comparison of | ||
| + | user-defined lesion patterns to automated/automatic lesion segmentation, | ||
| + | definition of comparison metrics. | ||
| + | |||
| + | |||
</div> | </div> | ||
| Line 28: | Line 60: | ||
<h1>Progress</h1> | <h1>Progress</h1> | ||
| + | ====Feb 2008 Updates==== | ||
| + | * Provided Killian with a two channel lesion segmentation performed with EMSegment. | ||
| + | * Waiting on EMSegment update to resolve errors relating to additional input channels and weighting by image. ( We need the third channel since we have flair. ) | ||
| + | * Waiting on Marcel to update and provide a copy of his lesion analysis tool. | ||
| + | |||
| + | |||
| + | ====Jan 2008 Project Week==== | ||
| + | * attended EM-segment breakout session | ||
| + | * attended Slicer Module breakout session | ||
| + | * attended Registration breakout | ||
| + | * attended Batchmake breakout | ||
| + | * met with Marcel to review his method and results of lesion analysis | ||
| + | * met with Brad and Kilian to work out EM-Segment issues | ||
| + | * successfully built Slicer3 from SVN on Mac G4/G5 OSX 10.4 platforms | ||
| + | ** we would like to figure out how to get it to buildon on Mac x86 OSX 10.5 platform | ||
| + | * successfully ran EM-Segment on tutorial data-set | ||
| + | * successfully ran EM-Segment on lupus and normal control cases | ||
| + | ** we have not yet gotten the segmentation of white matter into normal and lesion to work | ||
| + | * met with Vince Magnotta regarding BRAINS/ITK lesion segmentation method | ||
| + | ** we will have a modified method to try in about two weeks | ||
| + | * we plan to submit a paper to [http://miccai2008.rutgers.edu/ MICCAI 2008] | ||
====June 2007 Project Week==== | ====June 2007 Project Week==== | ||
| − | + | First meeting as a new DBP in lupus. | |
| + | We received hands on training on ITK/VTK, as well as, EM Segment. | ||
| + | We processed lupus case using EM Segment. | ||
====Jan 2007 Project Half Week==== | ====Jan 2007 Project Half Week==== | ||
| − | + | Jeremy attended this meeting in order to learn about NA-MIC | |
| − | |||
</div> | </div> | ||
| Line 43: | Line 97: | ||
===References=== | ===References=== | ||
| − | * | + | * |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 17:02, 6 February 2008
Home < 2008 Winter Project Week:Lesions Return to 2008_Winter_Project_Week |
|
Key Investigators
- MIND: Jeremy Bockolt, Mark Scully
- BWH: Sonia Pujol
- Utah: Marcel Prastawa
- Iowa: Vincent Magnotta
- Kitware: Brad Davis
Objective
Our objective is to make progress on the roadmap initiative of creating an end to end solution that provides a tutorial for NA-MIC kit users to perform lesion analyses in Slicer3.
Approach, Plan
For this week, we will attend and participate in the following training relevant to this DBP activities:
- EM Segmenter User Group
- Plugins for Slicer3
- DBP Engineers Lunch
- Registration Breakout
- Batchmake
Fully analyze 2 lupus subjects and 2 matched healthy normal volunteer data-sets collected:
- EM Segment
- BRAINS
- Prastawa/Gerig Lesion analysis method
We see our roadmap initiative breaking into the following stages:
a) Criteria and definition of what does an expert define and perceive as a lesion (like rules on multi-contrast MRI), creation of a document/catalogue that guides manual segmentation but also algorithm developers.
b) Specification of advanced software for user-guided efficient segmentation of lesions (e.g. 3D level set evolution etc., definition of MRI contrast to use to do that, etc.).
c) Towards approaches towards fully automatic lesion segmentation using multi-contrast MRI.,
d) Validateion/test framework for comparison of user-defined lesion patterns to automated/automatic lesion segmentation, definition of comparison metrics.
Progress
Feb 2008 Updates
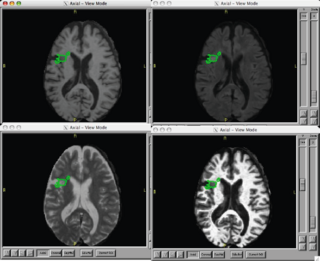
- Provided Killian with a two channel lesion segmentation performed with EMSegment.
- Waiting on EMSegment update to resolve errors relating to additional input channels and weighting by image. ( We need the third channel since we have flair. )
- Waiting on Marcel to update and provide a copy of his lesion analysis tool.
Jan 2008 Project Week
- attended EM-segment breakout session
- attended Slicer Module breakout session
- attended Registration breakout
- attended Batchmake breakout
- met with Marcel to review his method and results of lesion analysis
- met with Brad and Kilian to work out EM-Segment issues
- successfully built Slicer3 from SVN on Mac G4/G5 OSX 10.4 platforms
- we would like to figure out how to get it to buildon on Mac x86 OSX 10.5 platform
- successfully ran EM-Segment on tutorial data-set
- successfully ran EM-Segment on lupus and normal control cases
- we have not yet gotten the segmentation of white matter into normal and lesion to work
- met with Vince Magnotta regarding BRAINS/ITK lesion segmentation method
- we will have a modified method to try in about two weeks
- we plan to submit a paper to MICCAI 2008
June 2007 Project Week
First meeting as a new DBP in lupus. We received hands on training on ITK/VTK, as well as, EM Segment. We processed lupus case using EM Segment.
Jan 2007 Project Half Week
Jeremy attended this meeting in order to learn about NA-MIC