Difference between revisions of "2009 Summer Project Week TrigeminalNerve"
From NAMIC Wiki
MartaPeroni (talk | contribs) |
MartaPeroni (talk | contribs) |
||
| (16 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
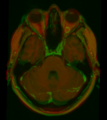
Image:Trigeminal_1.png|Overlay between warped atlas (red) and patient MRI (green) | Image:Trigeminal_1.png|Overlay between warped atlas (red) and patient MRI (green) | ||
Image:Trigeminal_2.png|Comparison between patient CT (gray), patient MRI (green) and warped atlas (red) | Image:Trigeminal_2.png|Comparison between patient CT (gray), patient MRI (green) and warped atlas (red) | ||
| + | Image:trigeminalUpdate2.jpg|result 1 | ||
| + | Image:trigeminalUpdate3.jpg|result 2 | ||
</gallery> | </gallery> | ||
| Line 16: | Line 18: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | We are tuning and testing multimodal image registration in [http://www.plastimatch.org plastimatch] to allow accurate localization of the trigeminal nerve. The final objective is to register an atlas to patient CT, in order to avoid 3T MRI. The registration would allow the physician to identify the trigeminal nerve on the atlas and plan the Cyberknife treatment accordingly. | + | We are tuning and testing multimodal image registration in [http://www.plastimatch.org plastimatch] to allow accurate localization of the trigeminal nerve. The final objective is to register an [http://www.bic.mni.mcgill.ca/brainweb/ atlas] to patient CT, in order to avoid 3T MRI. The registration would allow the physician to identify the trigeminal nerve on the atlas and plan the Cyberknife treatment accordingly. |
| Line 25: | Line 27: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | Our approach is to crop and " | + | Our approach is to crop and "adjust" the patient CT histogram and to crop the atlas to obtain a more robust result. The output of Bspline deformable registration is than compared to the patient 3T MRI (whenever possible in the same modality - T1). The MRIs are acquired generally with CISS sequence. |
Our plan for the project week is: | Our plan for the project week is: | ||
| − | * solve issues | + | * solve offset issues and parameter handling between rigid and non-rigid stage |
| − | * | + | * do we really need to "adjust" CT histogram? |
* investigate why the cropping is necessary for the registration to produce an output "acceptable" | * investigate why the cropping is necessary for the registration to produce an output "acceptable" | ||
| Line 37: | Line 39: | ||
<div style="width: 40%; float: left;"> | <div style="width: 40%; float: left;"> | ||
| − | <h3>Progress</h3> | + | <h3>Progress as on 06/26</h3> |
| − | + | * full pipeline implemented (coarse alignment + rigid + affine + deformable) | |
| + | * we need to cut-off the CT histogram or re-implement MI | ||
| + | * cropping the CT to the same region of the atlas is necessary | ||
| + | * new ideas: validation / correction of registration by mean of corresponding points | ||
| + | * still needs work: rigid stage needs to be optimized (or maybe not included?) | ||
| + | |||
| + | |||
</div> | </div> | ||
Latest revision as of 15:00, 26 June 2009
Home < 2009 Summer Project Week TrigeminalNerve
Key Investigators
- PoliMI: Marta Peroni
- UMG: Maria Francesca Spadea
- MGH: Greg Sharp
Objective
We are tuning and testing multimodal image registration in plastimatch to allow accurate localization of the trigeminal nerve. The final objective is to register an atlas to patient CT, in order to avoid 3T MRI. The registration would allow the physician to identify the trigeminal nerve on the atlas and plan the Cyberknife treatment accordingly.
Approach, Plan
Our approach is to crop and "adjust" the patient CT histogram and to crop the atlas to obtain a more robust result. The output of Bspline deformable registration is than compared to the patient 3T MRI (whenever possible in the same modality - T1). The MRIs are acquired generally with CISS sequence.
Our plan for the project week is:
- solve offset issues and parameter handling between rigid and non-rigid stage
- do we really need to "adjust" CT histogram?
- investigate why the cropping is necessary for the registration to produce an output "acceptable"
Progress as on 06/26
- full pipeline implemented (coarse alignment + rigid + affine + deformable)
- we need to cut-off the CT histogram or re-implement MI
- cropping the CT to the same region of the atlas is necessary
- new ideas: validation / correction of registration by mean of corresponding points
- still needs work: rigid stage needs to be optimized (or maybe not included?)
References
- Klein A, Andersson J, Ardekani BA et al., Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration., Neuroimage. 2009 Jul 1;46(3):786-802. Epub 2009 Jan 13.
- Chakravarty MM, Sadikot AF, Germann J, Bertrand G, Collins DL., Towards a validation of atlas warping techniques., Med Image Anal. 2008 Dec;12(6):713-26. Epub 2008 May 13.
- Borchers JD 3rd, Yang HJ, Sakamoto GT, Howes GA, Gupta G, Chang SD, Adler JR Jr., Cyberknife stereotactic radiosurgical rhizotomy for trigeminal neuralgia: anatomic and morphological considerations., Neurosurgery. 2009 Feb;64(2 Suppl):A91-5.
- Antypas C, Pantelis E., Performance evaluation of a CyberKnife G4 image-guided robotic stereotactic radiosurgery system. Phys Med Biol. 2008 Sep 7;53(17):4697-718. Epub 2008 Aug 11.